An integrated genomic approach for rapid delineation of candidate genes regulating agro-morphological traits in chickpea
- PMID: 25335477
- PMCID: PMC4263302
- DOI: 10.1093/dnares/dsu031
An integrated genomic approach for rapid delineation of candidate genes regulating agro-morphological traits in chickpea
Abstract
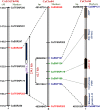
The identification and fine mapping of robust quantitative trait loci (QTLs)/genes governing important agro-morphological traits in chickpea still lacks systematic efforts at a genome-wide scale involving wild Cicer accessions. In this context, an 834 simple sequence repeat and single-nucleotide polymorphism marker-based high-density genetic linkage map between cultivated and wild parental accessions (Cicer arietinum desi cv. ICC 4958 and Cicer reticulatum wild cv. ICC 17160) was constructed. This inter-specific genetic map comprising eight linkage groups spanned a map length of 949.4 cM with an average inter-marker distance of 1.14 cM. Eleven novel major genomic regions harbouring 15 robust QTLs (15.6-39.8% R(2) at 4.2-15.7 logarithm of odds) associated with four agro-morphological traits (100-seed weight, pod and branch number/plant and plant hairiness) were identified and mapped on chickpea chromosomes. Most of these QTLs showed positive additive gene effects with effective allelic contribution from ICC 4958, particularly for increasing seed weight (SW) and pod and branch number. One robust SW-influencing major QTL region (qSW4.2) has been narrowed down by combining QTL mapping with high-resolution QTL region-specific association analysis, differential expression profiling and gene haplotype-based association/LD mapping. This enabled to delineate a strong SW-regulating ABI3VP1 transcription factor (TF) gene at trait-specific QTL interval and consequently identified favourable natural allelic variants and superior high seed weight-specific haplotypes in the upstream regulatory region of this gene showing increased transcript expression during seed development. The genes (TFs) harbouring diverse trait-regulating QTLs, once validated and fine-mapped by our developed rapid integrated genomic approach and through gene/QTL map-based cloning, can be utilized as potential candidates for marker-assisted genetic enhancement of chickpea.
Keywords: QTLs; SNP; SSR; chickpea; transcription factor; wild.
© The Author 2014. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures


References
-
- Zohary D., Hopf M. Domestication of plants in the old world. Oxford: Clarendon Press; 2000. Pulses; pp. 108–11.
-
- Muehlbauer F.J. Breeding for stress tolerance in cool season food legume. In: Singh K.B., Saxena M.C., editors. Use of wild species as a source of resistance in cool-season food legume crops. eds. Chichester, UK: Wiley; 1993. pp. 359–72.
-
- Berger J.D., Turner N.C. Chickpea breeding and management. In: Yadav S.S., Redden R.J., Chen W., Sharma B., editors. The ecology of chickpea. eds. UK: CAB International Publishing; 2007. pp. 47–71.
-
- Ladizinsky G., Adler A. Genetic relationships among the annual species of Cicer L. Theor. App. Genet. 1976a;48:197–203. - PubMed
-
- Ladizinsky G., Adler A. The origin of chickpea Cicer arietinum L. Euphytica. 1976b;25:211–17.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

