Integration host factor assembly at the cohesive end site of the bacteriophage lambda genome: implications for viral DNA packaging and bacterial gene regulation
- PMID: 25335823
- PMCID: PMC4263431
- DOI: 10.1021/bi501025s
Integration host factor assembly at the cohesive end site of the bacteriophage lambda genome: implications for viral DNA packaging and bacterial gene regulation
Abstract
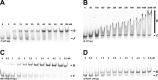
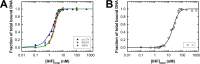
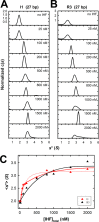
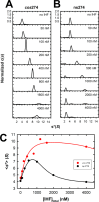
Integration host factor (IHF) is an Escherichia coli protein involved in (i) condensation of the bacterial nucleoid and (ii) regulation of a variety of cellular functions. In its regulatory role, IHF binds to a specific sequence to introduce a strong bend into the DNA; this provides a duplex architecture conducive to the assembly of site-specific nucleoprotein complexes. Alternatively, the protein can bind in a sequence-independent manner that weakly bends and wraps the duplex to promote nucleoid formation. IHF is also required for the development of several viruses, including bacteriophage lambda, where it promotes site-specific assembly of a genome packaging motor required for lytic development. Multiple IHF consensus sequences have been identified within the packaging initiation site (cos), and we here interrogate IHF-cos binding interactions using complementary electrophoretic mobility shift (EMS) and analytical ultracentrifugation (AUC) approaches. IHF recognizes a single consensus sequence within cos (I1) to afford a strongly bent nucleoprotein complex. In contrast, IHF binds weakly but with positive cooperativity to nonspecific DNA to afford an ensemble of complexes with increasing masses and levels of condensation. Global analysis of the EMS and AUC data provides constrained thermodynamic binding constants and nearest neighbor cooperativity factors for binding of IHF to I1 and to nonspecific DNA substrates. At elevated IHF concentrations, the nucleoprotein complexes undergo a transition from a condensed to an extended rodlike conformation; specific binding of IHF to I1 imparts a significant energy barrier to the transition. The results provide insight into how IHF can assemble specific regulatory complexes in the background of extensive nonspecific DNA condensation.
Figures



Similar articles
-
Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: implications for the assembly of a genome-packaging motor.Biochemistry. 2006 Apr 25;45(16):5180-9. doi: 10.1021/bi052284b. Biochemistry. 2006. PMID: 16618107
-
Physical and Functional Characterization of a Viral Genome Maturation Complex.Biophys J. 2017 Apr 25;112(8):1551-1560. doi: 10.1016/j.bpj.2017.02.041. Biophys J. 2017. PMID: 28445747 Free PMC article.
-
The interaction of Escherichia coli integration host factor with the cohesive end sites of phages lambda and 21.Nucleic Acids Res. 1988 Mar 25;16(5):2015-30. doi: 10.1093/nar/16.5.2015. Nucleic Acids Res. 1988. PMID: 2965807 Free PMC article.
-
Bending and supercoiling of DNA at the attachment site of bacteriophage lambda.Trends Biochem Sci. 1990 Jun;15(6):222-7. doi: 10.1016/0968-0004(90)90034-9. Trends Biochem Sci. 1990. PMID: 2166364 Review.
-
Interaction between bacteriophage lambda and its Escherichia coli host.Curr Opin Genet Dev. 1992 Oct;2(5):727-38. doi: 10.1016/s0959-437x(05)80133-9. Curr Opin Genet Dev. 1992. PMID: 1458022 Review.
Cited by
-
The Site-Specific Recombination System of the Escherichia coli Bacteriophage Φ24B.Front Microbiol. 2020 Oct 9;11:578056. doi: 10.3389/fmicb.2020.578056. eCollection 2020. Front Microbiol. 2020. PMID: 33162958 Free PMC article.
-
Bacteriophage lambda: Early pioneer and still relevant.Virology. 2015 May;479-480:310-30. doi: 10.1016/j.virol.2015.02.010. Epub 2015 Mar 3. Virology. 2015. PMID: 25742714 Free PMC article. Review.
-
Analytical Ultracentrifugation as a Tool to Study Nonspecific Protein-DNA Interactions.Methods Enzymol. 2015;562:305-30. doi: 10.1016/bs.mie.2015.04.009. Epub 2015 Jun 13. Methods Enzymol. 2015. PMID: 26412658 Free PMC article. Review.
-
The Characteristics and Genome Analysis of vB_AviM_AVP, the First Phage Infecting Aerococcus viridans.Viruses. 2019 Jan 26;11(2):104. doi: 10.3390/v11020104. Viruses. 2019. PMID: 30691182 Free PMC article.
-
Genomic and ecological study of two distinctive freshwater bacteriophages infecting a Comamonadaceae bacterium.Sci Rep. 2018 May 22;8(1):7989. doi: 10.1038/s41598-018-26363-y. Sci Rep. 2018. PMID: 29789681 Free PMC article.
References
-
- Azam T. A.; Ishihama A. (1999) Twelve Species of the Nucleoid-associated Protein from Escherichia coli: Sequence Recognition Specificity and DNA Binding Affinity. J. Biol. Chem. 274, 33105–33113. - PubMed
-
- Sarkar T.; Petrov A. S.; Vitko J. R.; Santai C. T.; Harvey S. C.; Mukerji I.; Hud N. V. (2009) Integration Host Factor (IHF) Dictates the Structure of Polyamine-DNA Condensates: Implications for the Role of IHF in the Compaction of Bacterial Chromatin. Biochemistry 48, 667–675. - PubMed
-
- Swinger K. K.; Rice P. A. (2004) IHF and HU: Flexible Architects of Bent DNA. Curr. Opin. Struct. Biol. 14, 28–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

