Assessing the role of a medication-indication resource in the treatment relation extraction from clinical text
- PMID: 25336593
- PMCID: PMC5901126
- DOI: 10.1136/amiajnl-2014-002954
Assessing the role of a medication-indication resource in the treatment relation extraction from clinical text
Abstract
Objective: To evaluate the contribution of the MEDication Indication (MEDI) resource and SemRep for identifying treatment relations in clinical text.
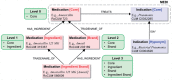
Materials and methods: We first processed clinical documents with SemRep to extract the Unified Medical Language System (UMLS) concepts and the treatment relations between them. Then, we incorporated MEDI into a simple algorithm that identifies treatment relations between two concepts if they match a medication-indication pair in this resource. For a better coverage, we expanded MEDI using ontology relationships from RxNorm and UMLS Metathesaurus. We also developed two ensemble methods, which combined the predictions of SemRep and the MEDI algorithm. We evaluated our selected methods on two datasets, a Vanderbilt corpus of 6864 discharge summaries and the 2010 Informatics for Integrating Biology and the Bedside (i2b2)/Veteran's Affairs (VA) challenge dataset.
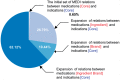
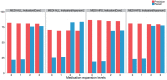
Results: The Vanderbilt dataset included 958 manually annotated treatment relations. A double annotation was performed on 25% of relations with high agreement (Cohen's κ = 0.86). The evaluation consisted of comparing the manual annotated relations with the relations identified by SemRep, the MEDI algorithm, and the two ensemble methods. On the first dataset, the best F1-measure results achieved by the MEDI algorithm and the union of the two resources (78.7 and 80, respectively) were significantly higher than the SemRep results (72.3). On the second dataset, the MEDI algorithm achieved better precision and significantly lower recall values than the best system in the i2b2 challenge. The two systems obtained comparable F1-measure values on the subset of i2b2 relations with both arguments in MEDI.
Conclusions: Both SemRep and MEDI can be used to extract treatment relations from clinical text. Knowledge-based extraction with MEDI outperformed use of SemRep alone, but superior performance was achieved by integrating both systems. The integration of knowledge-based resources such as MEDI into information extraction systems such as SemRep and the i2b2 relation extractors may improve treatment relation extraction from clinical text.
Keywords: MEDI; SemRep; natural language processing; treatment relation extraction.
© The Author 2014. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Cebul RD, Love TE, Jain AK, et al. Electronic health records and quality of diabetes care. N Engl J Med. 2011;365:825–33. - PubMed
-
- Roth CP, Lim YW, Pevnick JM, et al. The challenge of measuring quality of care from the electronic health record. Am J Med Qual. 2009;24:385–94. - PubMed
-
- Roth MT, Weinberger M, Campbell WH. Measuring the quality of medication use in older adults. J Am Geriatr Soc. 2009;57:1096–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

