The role of macrobiota in structuring microbial communities along rocky shores
- PMID: 25337459
- PMCID: PMC4203024
- DOI: 10.7717/peerj.631
The role of macrobiota in structuring microbial communities along rocky shores
Abstract
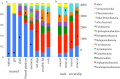
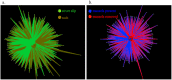
Rocky shore microbial diversity presents an excellent system to test for microbial habitat specificity or generality, enabling us to decipher how common macrobiota shape microbial community structure. At two coastal locations in the northeast Pacific Ocean, we show that microbial composition was significantly different between inert surfaces, the biogenic surfaces that included rocky shore animals and an alga, and the water column plankton. While all sampled entities had a core of common OTUs, rare OTUs drove differences among biotic and abiotic substrates. For the mussel Mytilus californianus, the shell surface harbored greater alpha diversity compared to internal tissues of the gill and siphon. Strikingly, a 7-year experimental removal of this mussel from tidepools did not significantly alter the microbial community structure of microbes associated with inert surfaces when compared with unmanipulated tidepools. However, bacterial taxa associated with nitrate reduction had greater relative abundance with mussels present, suggesting an impact of increased animal-derived nitrogen on a subset of microbial metabolism. Because the presence of mussels did not affect the structure and diversity of the microbial community on adjacent inert substrates, microbes in this rocky shore environment may be predominantly affected through direct physical association with macrobiota.
Keywords: 16S; Ammonium; Animal excretion; Host-microbe; Mytilus californianus; Nitrification; Nitrogen cycling; Rocky intertidal; Tatoosh Island; Tidepool.
Figures




References
-
- Amaral-Zettler L, Artigas LP, Baross J, Loka Bharathi PA, Boetius A, Chandramohan D, Herndl G, Kogure K, Neal P, Pedros-Alio C, Ramette A, Schouten S, Stal L, Thessen A, de Leeuw J, Sogin M. A global census of marine microbes. In: McIntyre A, editor. Life in the world’s oceans: diversity, distribution, and abundance. London: Blackwell Publishing; 2010. pp. 223–245.
-
- Barber RT, Smith RL. Coastal upwelling ecosystems. In: Longhurst A, editor. Analysis of marine ecosystems. New York: Academic Press; 1981. pp. 31–68.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

