DANN: a deep learning approach for annotating the pathogenicity of genetic variants
- PMID: 25338716
- PMCID: PMC4341060
- DOI: 10.1093/bioinformatics/btu703
DANN: a deep learning approach for annotating the pathogenicity of genetic variants
Abstract
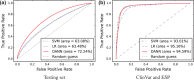
Annotating genetic variants, especially non-coding variants, for the purpose of identifying pathogenic variants remains a challenge. Combined annotation-dependent depletion (CADD) is an algorithm designed to annotate both coding and non-coding variants, and has been shown to outperform other annotation algorithms. CADD trains a linear kernel support vector machine (SVM) to differentiate evolutionarily derived, likely benign, alleles from simulated, likely deleterious, variants. However, SVMs cannot capture non-linear relationships among the features, which can limit performance. To address this issue, we have developed DANN. DANN uses the same feature set and training data as CADD to train a deep neural network (DNN). DNNs can capture non-linear relationships among features and are better suited than SVMs for problems with a large number of samples and features. We exploit Compute Unified Device Architecture-compatible graphics processing units and deep learning techniques such as dropout and momentum training to accelerate the DNN training. DANN achieves about a 19% relative reduction in the error rate and about a 14% relative increase in the area under the curve (AUC) metric over CADD's SVM methodology.
Availability and implementation: All data and source code are available at https://cbcl.ics.uci.edu/public_data/DANN/.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Baker M. (2012) One-stop shop for disease genes. Nature , 491, 171. - PubMed
-
- Franc V., Sonnenburg S. (2009) Optimized cutting plane algorithm for large-scale risk minimization. J. Mach. Learn. Res. , 10, 2157–2192.
-
- Pedregosa F., et al. . (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res. , 12, 2825–2830.

