Uneven genetic robustness of HIV-1 integrase
- PMID: 25339768
- PMCID: PMC4301135
- DOI: 10.1128/JVI.02451-14
Uneven genetic robustness of HIV-1 integrase
Abstract
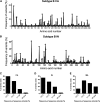
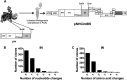
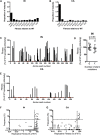
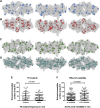
Genetic robustness (tolerance of mutation) may be a naturally selected property in some viruses, because it should enhance adaptability. Robustness should be especially beneficial to viruses like HIV-1 that exhibit high mutation rates and exist in immunologically hostile environments. Surprisingly, however, the HIV-1 capsid protein (CA) exhibits extreme fragility. To determine whether fragility is a general property of HIV-1 proteins, we created a large library of random, single-amino-acid mutants in HIV-1 integrase (IN), covering >40% of amino acid positions. Despite similar degrees of sequence variation in naturally occurring IN and CA sequences, we found that HIV-1 IN was significantly more robust than CA, with random nonsilent IN mutations only half as likely to cause lethal defects. Interestingly, IN and CA were similar in that a subset of mutations with high in vitro fitness were rare in natural populations. IN mutations of this type were more likely to occur in the buried interior of the modeled HIV-1 intasome, suggesting that even very subtle fitness effects suppress variation in natural HIV-1 populations. Lethal mutations, in particular those that perturbed particle production, proteolytic processing, and particle-associated IN levels, were strikingly localized at specific IN subunit interfaces. This observation strongly suggests that binding interactions between particular IN subunits regulate proteolysis during HIV-1 virion morphogenesis. Overall, use of the IN mutant library in conjunction with structural models demonstrates the overall robustness of IN and highlights particular regions of vulnerability that may be targeted in therapeutic interventions.
Importance: The HIV-1 integrase (IN) protein is responsible for the integration of the viral genome into the host cell chromosome. To measure the capacity of IN to maintain function in the face of mutation, and to probe structure/function relationships, we created a library of random single-amino-acid IN mutations that could mimic the types of mutations that naturally occur during HIV-1 infection. Previously, we measured the robustness of HIV-1 capsid in this manner and determined that it is extremely intolerant of mutation. In contrast to CA, HIV-1 IN proved relatively robust, with far fewer mutations causing lethal defects. However, when we subsequently mapped the lethal mutations onto a model of the structure of the multisubunit IN-viral DNA complex, we found the lethal mutations that caused virus morphogenesis defects tended to be highly localized at subunit interfaces. This discovery of vulnerable regions of HIV-1 IN could inform development of novel therapeutics.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Extreme genetic fragility of the HIV-1 capsid.PLoS Pathog. 2013;9(6):e1003461. doi: 10.1371/journal.ppat.1003461. Epub 2013 Jun 20. PLoS Pathog. 2013. PMID: 23818857 Free PMC article.
-
Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells.J Virol. 2015 Jan;89(1):643-51. doi: 10.1128/JVI.03043-14. Epub 2014 Oct 22. J Virol. 2015. PMID: 25339776 Free PMC article.
-
Compensatory substitutions in the HIV-1 capsid reduce the fitness cost associated with resistance to a capsid-targeting small-molecule inhibitor.J Virol. 2015 Jan;89(1):208-19. doi: 10.1128/JVI.01411-14. Epub 2014 Oct 15. J Virol. 2015. PMID: 25320302 Free PMC article.
-
Going beyond Integration: The Emerging Role of HIV-1 Integrase in Virion Morphogenesis.Viruses. 2020 Sep 9;12(9):1005. doi: 10.3390/v12091005. Viruses. 2020. PMID: 32916894 Free PMC article. Review.
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
Cited by
-
In vivo mutation rates and the landscape of fitness costs of HIV-1.Virus Evol. 2017 Mar 2;3(1):vex003. doi: 10.1093/ve/vex003. eCollection 2017 Jan. Virus Evol. 2017. PMID: 28458914 Free PMC article.
-
Vulnerable targets in HIV-1 Pol for attenuation-based vaccine design.Virology. 2021 Feb;554:1-8. doi: 10.1016/j.virol.2020.12.003. Epub 2020 Dec 10. Virology. 2021. PMID: 33316731 Free PMC article.
-
HIV-1 Integrase Binds the Viral RNA Genome and Is Essential during Virion Morphogenesis.Cell. 2016 Aug 25;166(5):1257-1268.e12. doi: 10.1016/j.cell.2016.07.044. Cell. 2016. PMID: 27565348 Free PMC article.
-
HIV-1 Protease, Reverse Transcriptase, and Integrase Variation.J Virol. 2016 Jun 10;90(13):6058-6070. doi: 10.1128/JVI.00495-16. Print 2016 Jul 1. J Virol. 2016. PMID: 27099321 Free PMC article.
-
A Highly Conserved Residue in HIV-1 Nef Alpha Helix 2 Modulates Protein Expression.mSphere. 2016 Nov 9;1(6):e00288-16. doi: 10.1128/mSphere.00288-16. eCollection 2016 Nov-Dec. mSphere. 2016. PMID: 27840851 Free PMC article.
References
-
- de Visser JA, Hermisson J, Wagner GP, Ancel Meyers L, Bagheri-Chaichian H, Blanchard JL, Chao L, Cheverud JM, Elena SF, Fontana W, Gibson G, Hansen TF, Krakauer D, Lewontin RC, Ofria C, Rice SH, von Dassow G, Wagner A, Whitlock MC. 2003. Perspective: evolution and detection of genetic robustness. Evolution 57:1959–1972. doi:10.1111/j.0014-3820.2003.tb00377.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

