The involvement of J-protein AtDjC17 in root development in Arabidopsis
- PMID: 25339971
- PMCID: PMC4189540
- DOI: 10.3389/fpls.2014.00532
The involvement of J-protein AtDjC17 in root development in Arabidopsis
Abstract
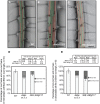
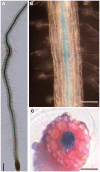
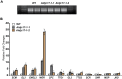
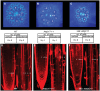
In a screen for root hair morphogenesis mutants in Arabidopsis thaliana L. we identified a T-DNA insertion within a type III J-protein AtDjC17 caused altered root hair development and reduced hair length. Root hairs were observed to develop from trichoblast and atrichoblast cell files in both Atdjc17 and 35S::AtDJC17. Localization of gene expression in the root using transgenic plants expressing proAtDjC17::GUS revealed constitutive expression in stele cells. No AtDJC17 expression was observed in epidermal, endodermal, or cortical layers. To explore the contrast between gene expression in the stele and epidermal phenotype, hand cut transverse sections of Atdjc17 roots were examined showing that the endodermal and cortical cell layers displayed increased anticlinal cell divisions. Aberrant cortical cell division in Atdjc17 is proposed as causal in ectopic root hair formation via the positional cue requirement that exists between cortical and epidermal cell in hair cell fate determination. Results indicate a requirement for AtDJC17 in position-dependent cell fate determination and illustrate an intriguing requirement for molecular co-chaperone activity during root development.
Keywords: J-family; J-proteins; development; heat shock protein; root hair; root patterning.
Figures


Similar articles
-
Positional signaling and expression of ENHANCER OF TRY AND CPC1 are tuned to increase root hair density in response to phosphate deficiency in Arabidopsis thaliana.PLoS One. 2013 Oct 9;8(10):e75452. doi: 10.1371/journal.pone.0075452. eCollection 2013. PLoS One. 2013. PMID: 24130712 Free PMC article.
-
Cell Fate Determination and the Switch from Diffuse Growth to Planar Polarity in Arabidopsis Root Epidermal Cells.Front Plant Sci. 2015 Dec 23;6:1163. doi: 10.3389/fpls.2015.01163. eCollection 2015. Front Plant Sci. 2015. PMID: 26779192 Free PMC article. Review.
-
The Density and Length of Root Hairs Are Enhanced in Response to Cadmium and Arsenic by Modulating Gene Expressions Involved in Fate Determination and Morphogenesis of Root Hairs in Arabidopsis.Front Plant Sci. 2016 Nov 23;7:1763. doi: 10.3389/fpls.2016.01763. eCollection 2016. Front Plant Sci. 2016. PMID: 27933081 Free PMC article.
-
The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root.Dev Biol. 1994 Dec;166(2):740-54. doi: 10.1006/dbio.1994.1352. Dev Biol. 1994. PMID: 7813791
-
[The mechanism of root hair development and molecular regulation in plants].Yi Chuan. 2007 Apr;29(4):413-9. doi: 10.1360/yc-007-0413. Yi Chuan. 2007. PMID: 17548302 Review. Chinese.
Cited by
-
GmDNJ1, a type-I heat shock protein 40 (HSP40), is responsible for both Growth and heat tolerance in soybean.Plant Direct. 2021 Jan 25;5(1):e00298. doi: 10.1002/pld3.298. eCollection 2021 Jan. Plant Direct. 2021. PMID: 33532690 Free PMC article.
-
The expanding world of plant J-domain proteins.CRC Crit Rev Plant Sci. 2019;38(5-6):382-400. doi: 10.1080/07352689.2019.1693716. Epub 2019 Dec 10. CRC Crit Rev Plant Sci. 2019. PMID: 33223602 Free PMC article.
-
Roles and maturation of iron-sulfur proteins in plastids.J Biol Inorg Chem. 2018 Jun;23(4):545-566. doi: 10.1007/s00775-018-1532-1. Epub 2018 Jan 18. J Biol Inorg Chem. 2018. PMID: 29349662 Free PMC article. Review.
-
Analysis of the impact of indole-3-acetic acid (IAA) on gene expression during leaf senescence in Arabidopsis thaliana.Physiol Mol Biol Plants. 2020 Apr;26(4):733-745. doi: 10.1007/s12298-019-00752-7. Epub 2020 Feb 6. Physiol Mol Biol Plants. 2020. PMID: 32255936 Free PMC article.
-
ArHsp40, a type 1 J-domain protein, is developmentally regulated and stress inducible in post-diapause Artemia franciscana.Cell Stress Chaperones. 2016 Nov;21(6):1077-1088. doi: 10.1007/s12192-016-0732-2. Epub 2016 Aug 31. Cell Stress Chaperones. 2016. PMID: 27581971 Free PMC article.
References
-
- Al-Whaibi M. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ. Sci. 23 139–150 10.1016/j.jksus.2010.06.022 - DOI
-
- Bekh-Ochir D., Shimada S., Yamagami A., Kanda S., Ogawa K., Nakazawa M., et al. (2013). A novel mitochondrial DnaJ/Hsp40 family protein BIL2 promotes plant growth and resistance against environmental stress in brassinosteroid signaling. Planta 237 1509–1525 10.1007/s00425-013-1859-3 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

