Nuclear and mitochondrial tRNA-lookalikes in the human genome
- PMID: 25339973
- PMCID: PMC4189335
- DOI: 10.3389/fgene.2014.00344
Nuclear and mitochondrial tRNA-lookalikes in the human genome
Abstract
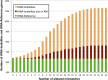
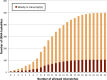
We are interested in identifying and characterizing loci of the human genome that harbor sequences resembling known mitochondrial and nuclear tRNAs. To this end, we used the known nuclear and mitochondrial tRNA genes (the "tRNA-Reference" set) to search for "tRNA-lookalikes" and found many such loci at different levels of sequence conservation. We find that the large majority of these tRNA-lookalikes resemble mitochondrial tRNAs and exhibit a skewed over-representation in favor of some mitochondrial anticodons. Our analysis shows that the tRNA-lookalikes have infiltrated specific chromosomes and are preferentially located in close proximity to known nuclear tRNAs (z-score ≤ -2.54, P-value ≤ 0.00394). Examination of the transcriptional potential of these tRNA-lookalike loci using public transcript annotations revealed that more than 20% of the lookalikes are transcribed as part of either known protein-coding pre-mRNAs, known lncRNAs, or known non-protein-coding RNAs, while public RNA-seq data perfectly agreed with the endpoints of tRNA-lookalikes. Interestingly, we found that tRNA-lookalikes are significantly depleted in known genetic variations associated with human health and disease whereas the known tRNAs are enriched in such variations. Lastly, a manual comparative analysis of the cloverleaf structure of several of the transcribed tRNA-lookalikes revealed no disruptive mutations suggesting the possibility that these loci give rise to functioning tRNA molecules.
Keywords: human genome; mitochondrial tRNA; nuclear tRNA; tRNA; tRNA fragment.
Figures


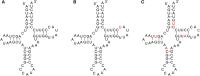

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

