Interplay of microRNA and epigenetic regulation in the human regulatory network
- PMID: 25339974
- PMCID: PMC4186481
- DOI: 10.3389/fgene.2014.00345
Interplay of microRNA and epigenetic regulation in the human regulatory network
Abstract
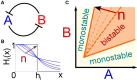
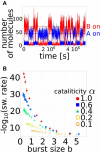
The expression of protein-coding genes is controlled by a complex network of regulatory interactions. It is becoming increasingly appreciated that post-transcriptional repression by microRNAs, a class of small non-coding RNAs, is a key layer of regulation in several biological processes. In this contribution, we discuss the interplay between microRNAs and epigenetic regulators. Among the mixed genetic circuits composed by these two different kinds of regulation, it seems that a central role is played by double-negative feedback loops in which a microRNA inhibits an epigenetic regulator and in turn is controlled at the epigenetic level by the same regulator. We discuss a few relevant properties of this class of network motifs and their potential role in cell differentiation. In particular, using mathematical modeling we show how this particular circuit can exhibit a switch-like behavior between two alternative steady states, while being robust to stochastic transitions between these two states, a feature presumably required for circuits involved in cell fate decision. Finally, we present a list of putative double-negative feedback loops from a literature survey combined with bioinformatic analysis, and discuss in detail a few examples.
Keywords: epigenetic regulation; feedback loops; microRNAs; network motifs.
Figures

References
-
- Alon U. (2006). An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL: Chapman and Hall/ CRC Press
LinkOut - more resources
Full Text Sources
Other Literature Sources

