A taxonomy of bacterial microcompartment loci constructed by a novel scoring method
- PMID: 25340524
- PMCID: PMC4207490
- DOI: 10.1371/journal.pcbi.1003898
A taxonomy of bacterial microcompartment loci constructed by a novel scoring method
Abstract
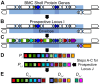
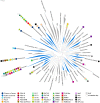
Bacterial microcompartments (BMCs) are proteinaceous organelles involved in both autotrophic and heterotrophic metabolism. All BMCs share homologous shell proteins but differ in their complement of enzymes; these are typically encoded adjacent to shell protein genes in genetic loci, or operons. To enable the identification and prediction of functional (sub)types of BMCs, we developed LoClass, an algorithm that finds putative BMC loci and inventories, weights, and compares their constituent pfam domains to construct a locus similarity network and predict locus (sub)types. In addition to using LoClass to analyze sequences in the Non-redundant Protein Database, we compared predicted BMC loci found in seven candidate bacterial phyla (six from single-cell genomic studies) to the LoClass taxonomy. Together, these analyses resulted in the identification of 23 different types of BMCs encoded in 30 distinct locus (sub)types found in 23 bacterial phyla. These include the two carboxysome types and a divergent set of metabolosomes, BMCs that share a common catalytic core and process distinct substrates via specific signature enzymes. Furthermore, many Candidate BMCs were found that lack one or more core metabolosome components, including one that is predicted to represent an entirely new paradigm for BMC-associated metabolism, joining the carboxysome and metabolosome. By placing these results in a phylogenetic context, we provide a framework for understanding the horizontal transfer of these loci, a starting point for studies aimed at understanding the evolution of BMCs. This comprehensive taxonomy of BMC loci, based on their constituent protein domains, foregrounds the functional diversity of BMCs and provides a reference for interpreting the role of BMC gene clusters encoded in isolate, single cell, and metagenomic data. Many loci encode ancillary functions such as transporters or genes for cofactor assembly; this expanded vocabulary of BMC-related functions should be useful for design of genetic modules for introducing BMCs in bioengineering applications.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

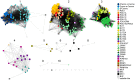


References
-
- Van Niftrik L, Jetten MSM (2012) Anaerobic ammonium-oxidizing bacteria: unique microorganisms with exceptional properties. Microbiol Mol Biol Rev 76: 585–596 Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3429623/. Accessed 23 May 2014. - PMC - PubMed
-
- Cornejo E, Abreu N, Komeili A (2014) Compartmentalization and organelle formation in bacteria. Curr Opin Cell Biol 26: 132–138 Available: http://www.ncbi.nlm.nih.gov/pubmed/24440431. Accessed 23 May 2014. - PMC - PubMed
-
- Kerfeld CA, Heinhorst S, Cannon GC (2010) Bacterial microcompartments. Annu Rev Microbiol 64: 391–408 Available: http://www.ncbi.nlm.nih.gov/pubmed/20825353. Accessed 29 April 2014. - PubMed
-
- Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA (2008) Bacterial microcompartments: their properties and paradoxes. Bioessays 30: 1084–1095 Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3272490/. Accessed 23 May 2014. - PMC - PubMed
-
- Yeates TO, Jorda J, Bobik TA (2013) The shells of BMC-type microcompartment organelles in bacteria. J Mol Microbiol Biotechnol 23: 290–299 Available: http://www.karger.com/Article/FullText/351347. Accessed 23 May 2014. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

