Topoisomerase II is required for the proper separation of heterochromatic regions during Drosophila melanogaster female meiosis
- PMID: 25340780
- PMCID: PMC4207608
- DOI: 10.1371/journal.pgen.1004650
Topoisomerase II is required for the proper separation of heterochromatic regions during Drosophila melanogaster female meiosis
Abstract
Heterochromatic homology ensures the segregation of achiasmate chromosomes during meiosis I in Drosophila melanogaster females, perhaps as a consequence of the heterochromatic threads that connect achiasmate homologs during prometaphase I. Here, we ask how these threads, and other possible heterochromatic entanglements, are resolved prior to anaphase I. We show that the knockdown of Topoisomerase II (Top2) by RNAi in the later stages of meiosis results in a specific defect in the separation of heterochromatic regions after spindle assembly. In Top2 RNAi-expressing oocytes, heterochromatic regions of both achiasmate and chiasmate chromosomes often failed to separate during prometaphase I and metaphase I. Heterochromatic regions were stretched into long, abnormal projections with centromeres localizing near the tips of the projections in some oocytes. Despite these anomalies, we observed bipolar spindles in most Top2 RNAi-expressing oocytes, although the obligately achiasmate 4th chromosomes exhibited a near complete failure to move toward the spindle poles during prometaphase I. Both achiasmate and chiasmate chromosomes displayed defects in biorientation. Given that euchromatic regions separate much earlier in prophase, no defects were expected or observed in the ability of euchromatic regions to separate during late prophase upon knockdown of Top2 at mid-prophase. Finally, embryos from Top2 RNAi-expressing females frequently failed to initiate mitotic divisions. These data suggest both that Topoisomerase II is involved in the resolution of heterochromatic DNA entanglements during meiosis I and that these entanglements must be resolved in order to complete meiosis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

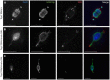
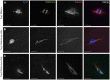
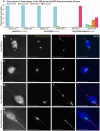
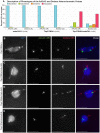
Similar articles
-
Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes.PLoS Genet. 2009 Jan;5(1):e1000348. doi: 10.1371/journal.pgen.1000348. Epub 2009 Jan 23. PLoS Genet. 2009. PMID: 19165317 Free PMC article.
-
There are two mechanisms of achiasmate segregation in Drosophila females, one of which requires heterochromatic homology.Dev Genet. 1992;13(6):440-67. doi: 10.1002/dvg.1020130608. Dev Genet. 1992. PMID: 1304424
-
Heterochromatin-Associated Proteins HP1a and Piwi Collaborate to Maintain the Association of Achiasmate Homologs in Drosophila Oocytes.Genetics. 2016 May;203(1):173-89. doi: 10.1534/genetics.115.186460. Epub 2016 Mar 16. Genetics. 2016. PMID: 26984058 Free PMC article.
-
Requiem for distributive segregation: achiasmate segregation in Drosophila females.Trends Genet. 1993 Sep;9(9):310-7. doi: 10.1016/0168-9525(93)90249-h. Trends Genet. 1993. PMID: 8236460 Review.
-
A Brief History of Drosophila (Female) Meiosis.Genes (Basel). 2022 Apr 27;13(5):775. doi: 10.3390/genes13050775. Genes (Basel). 2022. PMID: 35627159 Free PMC article. Review.
Cited by
-
Topoisomerases Modulate the Timing of Meiotic DNA Breakage and Chromosome Morphogenesis in Saccharomyces cerevisiae.Genetics. 2020 May;215(1):59-73. doi: 10.1534/genetics.120.303060. Epub 2020 Mar 9. Genetics. 2020. PMID: 32152049 Free PMC article.
-
Achiasmy: Male Fruit Flies Are Not Ready to Mix.Front Cell Dev Biol. 2016 Jul 19;4:75. doi: 10.3389/fcell.2016.00075. eCollection 2016. Front Cell Dev Biol. 2016. PMID: 27486580 Free PMC article.
-
TOP-2 is differentially required for the proper maintenance of the cohesin subunit REC-8 on meiotic chromosomes in Caenorhabditis elegans spermatogenesis and oogenesis.Genetics. 2022 Sep 30;222(2):iyac120. doi: 10.1093/genetics/iyac120. Genetics. 2022. PMID: 35951744 Free PMC article.
-
Roles of Topoisomerases in Heterochromatin, Aging, and Diseases.Genes (Basel). 2019 Nov 1;10(11):884. doi: 10.3390/genes10110884. Genes (Basel). 2019. PMID: 31683993 Free PMC article. Review.
-
Identification of Suppressors of top-2 Embryonic Lethality in Caenorhabditis elegans.G3 (Bethesda). 2020 Apr 9;10(4):1183-1191. doi: 10.1534/g3.119.400927. G3 (Bethesda). 2020. PMID: 32086248 Free PMC article.
References
-
- Ashburner M, Golic KG, Hawley RS (2005) Female meiosis. Drosophila A Laboratory Handbook. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. pp.757–825.
-
- Hawley RS, Theurkauf WE (1993) Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet 9: 310–317. - PubMed
-
- Hawley RS, Irick H, Zitron AE, Haddox DA, Lohe A, et al. (1992) There are two mechanisms of achiasmate segregation in Drosophila females, one of which requires heterochromatic homology. Dev Genet 13: 440–467. - PubMed
-
- Karpen GH, Le MH, Le H (1996) Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science 273: 118–122. - PubMed
-
- Dernburg AF, Sedat JW, Hawley RS (1996) Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135–146. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

