Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation
- PMID: 25341086
- PMCID: PMC4333581
- DOI: 10.1021/ja5055862
Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation
Abstract
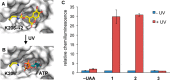
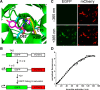
The site-specific incorporation of three new coumarin lysine analogues into proteins was achieved in bacterial and mammalian cells using an engineered pyrrolysyl-tRNA synthetase system. The genetically encoded coumarin lysines were successfully applied as fluorescent cellular probes for protein localization and for the optical activation of protein function. As a proof-of-principle, photoregulation of firefly luciferase was achieved in live cells by caging a key lysine residue, and excellent OFF to ON light-switching ratios were observed. Furthermore, two-photon and single-photon optochemical control of EGFP maturation was demonstrated, enabling the use of different, potentially orthogonal excitation wavelengths (365, 405, and 760 nm) for the sequential activation of protein function in live cells. These results demonstrate that coumarin lysines are a new and valuable class of optical probes that can be used for the investigation and regulation of protein structure, dynamics, function, and localization in live cells. The small size of coumarin, the site-specific incorporation, the application as both a light-activated caging group and as a fluorescent probe, and the broad range of excitation wavelengths are advantageous over other genetically encoded photocontrol systems and provide a precise and multifunctional tool for cellular biology.
Figures



References
-
- Bort G.; Gallavardin T.; Ogden D.; Dalko P. I. Angew. Chem., Int. Ed. 2013, 52, 4526–4537. - PubMed
-
- Krueger A. T.; Imperiali B. ChemBioChem 2013, 14, 788–799. - PubMed
-
- Goncalves M. S. Chem. Rev. 2009, 109, 190–212. - PubMed
-
- Brieke C.; Rohrbach F.; Gottschalk A.; Mayer G.; Heckel A. Angew. Chem., Int. Ed. 2012, 51, 8446–8476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

