Dendritic cells control fibroblastic reticular network tension and lymph node expansion
- PMID: 25341788
- PMCID: PMC4235005
- DOI: 10.1038/nature13814
Dendritic cells control fibroblastic reticular network tension and lymph node expansion
Abstract
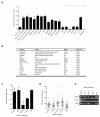
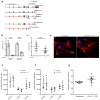
After immunogenic challenge, infiltrating and dividing lymphocytes markedly increase lymph node cellularity, leading to organ expansion. Here we report that the physical elasticity of lymph nodes is maintained in part by podoplanin (PDPN) signalling in stromal fibroblastic reticular cells (FRCs) and its modulation by CLEC-2 expressed on dendritic cells. We show in mouse cells that PDPN induces actomyosin contractility in FRCs via activation of RhoA/C and downstream Rho-associated protein kinase (ROCK). Engagement by CLEC-2 causes PDPN clustering and rapidly uncouples PDPN from RhoA/C activation, relaxing the actomyosin cytoskeleton and permitting FRC stretching. Notably, administration of CLEC-2 protein to immunized mice augments lymph node expansion. In contrast, lymph node expansion is significantly constrained in mice selectively lacking CLEC-2 expression in dendritic cells. Thus, the same dendritic cells that initiate immunity by presenting antigens to T lymphocytes also initiate remodelling of lymph nodes by delivering CLEC-2 to FRCs. CLEC-2 modulation of PDPN signalling permits FRC network stretching and allows for the rapid lymph node expansion--driven by lymphocyte influx and proliferation--that is the critical hallmark of adaptive immunity.
Figures




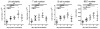



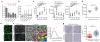
Comment in
-
Physiology: Relax and come in.Nature. 2014 Oct 23;514(7523):441-2. doi: 10.1038/514441a. Nature. 2014. PMID: 25341781 No abstract available.
References
-
- Girard J-P, Moussion C, Förster R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat Rev Immunol. 2012;12:762–773. - PubMed
-
- Junt T, Scandella E, Ludewig B. Form follows function: lymphoid tissue microarchitecture in antimicrobial immune defence. Nat Rev Immunol. 2008;8:764–775. - PubMed
-
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. - PubMed
-
- Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat. Immunol. 2007;8:1255–1265. - PubMed
-
- Katakai T. A novel reticular stromal structure in lymph node cortex: an immuno-platform for interactions among dendritic cells, T cells and B cells. International Immunology. 2004;16:1133–1142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

