Enzymatic DNA oxidation: mechanisms and biological significance
- PMID: 25341925
- PMCID: PMC4281339
- DOI: 10.5483/bmbrep.2014.47.11.223
Enzymatic DNA oxidation: mechanisms and biological significance
Abstract
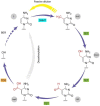
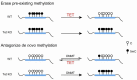
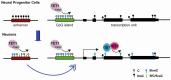
DNA methylation at cytosines (5mC) is a major epigenetic modification involved in the regulation of multiple biological processes in mammals. How methylation is reversed was until recently poorly understood. The family of dioxygenases commonly known as Ten-eleven translocation (Tet) proteins are responsible for the oxidation of 5mC into three new forms, 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Current models link Tet-mediated 5mC oxidation with active DNA demethylation. The higher oxidation products (5fC and 5caC) are recognized and excised by the DNA glycosylase TDG via the base excision repair pathway. Like DNA methyltransferases, Tet enzymes are important for embryonic development. We will examine the mechanism and biological significance of Tet-mediated 5mC oxidation in the context of pronuclear DNA demethylation in mouse early embryos. In contrast to its role in active demethylation in the germ cells and early embryo, a number of lines of evidence suggest that the intragenic 5hmC present in brain may act as a stable mark instead. This short review explores mechanistic aspects of TET oxidation activity, the impact Tet enzymes have on epigenome organization and their contribution to the regulation of early embryonic and neuronal development.
Figures
Similar articles
-
Structure and Function of TET Enzymes.Adv Exp Med Biol. 2022;1389:239-267. doi: 10.1007/978-3-031-11454-0_10. Adv Exp Med Biol. 2022. PMID: 36350513
-
Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA.Science. 2011 Sep 2;333(6047):1303-7. doi: 10.1126/science.1210944. Epub 2011 Aug 4. Science. 2011. PMID: 21817016 Free PMC article.
-
Direct decarboxylation of ten-eleven translocation-produced 5-carboxylcytosine in mammalian genomes forms a new mechanism for active DNA demethylation.Chem Sci. 2021 Jul 21;12(34):11322-11329. doi: 10.1039/d1sc02161c. eCollection 2021 Sep 1. Chem Sci. 2021. PMID: 34567494 Free PMC article.
-
TET-mediated active DNA demethylation: mechanism, function and beyond.Nat Rev Genet. 2017 Sep;18(9):517-534. doi: 10.1038/nrg.2017.33. Epub 2017 May 30. Nat Rev Genet. 2017. PMID: 28555658 Review.
-
Structure and Function of TET Enzymes.Adv Exp Med Biol. 2016;945:275-302. doi: 10.1007/978-3-319-43624-1_12. Adv Exp Med Biol. 2016. PMID: 27826843 Review.
Cited by
-
Structure and Function of TET Enzymes.Adv Exp Med Biol. 2022;1389:239-267. doi: 10.1007/978-3-031-11454-0_10. Adv Exp Med Biol. 2022. PMID: 36350513
-
Cell-Wide DNA De-Methylation and Re-Methylation of Purkinje Neurons in the Developing Cerebellum.PLoS One. 2016 Sep 1;11(9):e0162063. doi: 10.1371/journal.pone.0162063. eCollection 2016. PLoS One. 2016. PMID: 27583369 Free PMC article.
-
Gadd45a promotes DNA demethylation through TDG.Nucleic Acids Res. 2015 Apr 30;43(8):3986-97. doi: 10.1093/nar/gkv283. Epub 2015 Apr 6. Nucleic Acids Res. 2015. PMID: 25845601 Free PMC article.
-
Uracil-DNA Glycosylase UNG Promotes Tet-mediated DNA Demethylation.J Biol Chem. 2016 Jan 8;291(2):731-8. doi: 10.1074/jbc.M115.693861. Epub 2015 Nov 30. J Biol Chem. 2016. PMID: 26620559 Free PMC article.
-
DNA Hydroxymethylation at the Interface of the Environment and Nonalcoholic Fatty Liver Disease.Int J Environ Res Public Health. 2019 Aug 5;16(15):2791. doi: 10.3390/ijerph16152791. Int J Environ Res Public Health. 2019. PMID: 31387232 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

