Evolutionary history of Methyltransferase 1 genes in hexaploid wheat
- PMID: 25342325
- PMCID: PMC4223845
- DOI: 10.1186/1471-2164-15-922
Evolutionary history of Methyltransferase 1 genes in hexaploid wheat
Abstract
Background: Plant and animal methyltransferases are key enzymes involved in DNA methylation at cytosine residues, required for gene expression control and genome stability. Taking advantage of the new sequence surveys of the wheat genome recently released by the International Wheat Genome Sequencing Consortium, we identified and characterized MET1 genes in the hexaploid wheat Triticum aestivum (TaMET1).
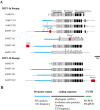
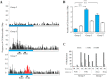
Results: Nine TaMET1 genes were identified and mapped on homoeologous chromosome groups 2A/2B/2D, 5A/5B/5D and 7A/7B/7D. Synteny analysis and evolution rates suggest that the genome organization of TaMET1 genes results from a whole genome duplication shared within the grass family, and a second gene duplication, which occurred specifically in the Triticeae tribe prior to the speciation of diploid wheat. Higher expression levels were observed for TaMET1 homoeologous group 2 genes compared to group 5 and 7, indicating that group 2 homoeologous genes are predominant at the transcriptional level, while group 5 evolved into pseudogenes. We show the connection between low expression levels, elevated evolution rates and unexpected enrichment in CG-dinucleotides (CG-rich isochores) at putative promoter regions of homoeologous group 5 and 7, but not of group 2 TaMET1 genes. Bisulfite sequencing reveals that these CG-rich isochores are highly methylated in a CG context, which is the expected target of TaMET1.
Conclusions: We retraced the evolutionary history of MET1 genes in wheat, explaining the predominance of group 2 homoeologous genes and suggest CG-DNA methylation as one of the mechanisms involved in wheat genome dynamics.
Figures




References
-
- Gaut BS. Evolutionary dynamics of grass genomes. New Phytol. 2002;154:15–28. doi: 10.1046/j.1469-8137.2002.00352.x. - DOI
-
- Feldman M, Lupton F, Miller T. Wheats. In: Smartt J, Simmonds N, editors. Evol Crops Ed 2 Longman Sci Lond. 1995. pp. 184–192.
-
- Yu J, Hu S, Wang J, Wong GK-S, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

