Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration
- PMID: 25342562
- PMCID: PMC4650581
- DOI: 10.1038/cr.2014.135
Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration
Abstract
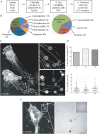
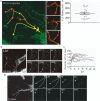
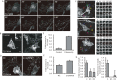
Cells communicate with each other through secreting and releasing proteins and vesicles. Many cells can migrate. In this study, we report the discovery of migracytosis, a cell migration-dependent mechanism for releasing cellular contents, and migrasomes, the vesicular structures that mediate migracytosis. As migrating cells move, they leave long tubular strands, called retraction fibers, behind them. Large vesicles, which contain numerous smaller vesicles, grow on the tips and intersections of retraction fibers. These fibers, which connect the vesicles with the main cell body, eventually break, and the vesicles are released into the extracellular space or directly taken up by surrounding cells. Since the formation of these vesicles is migration-dependent, we named them "migrasomes". We also found that cytosolic contents can be transported into migrasomes and released from the cell through migrasomes. We named this migration-dependent release mechanism "migracytosis".
Figures





Comment in
-
Migrasomes: a new organelle of migrating cells.Cell Res. 2015 Jan;25(1):1-2. doi: 10.1038/cr.2014.146. Epub 2014 Nov 7. Cell Res. 2015. PMID: 25378181 Free PMC article.
References
-
- 2Nagasawa J, Douglas WW, Schulz RA. Ultrastructural evidence of secretion by exocytosis and of synaptic-vesicle formation in posterior pituitary glands. Nature 1970; 227:407–409. - PubMed
-
- 3Trams EG, Lauter CJ, Salem N, Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 1981; 645:63–70. - PubMed
-
- 4Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. vesicle formation during reticulocyte maturation - association of plasma-membrane activities with released vesicles (exosomes). J Biol Chem 1987; 262:9412–9420. - PubMed
-
- 5Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014; 29:116–125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

