Dissection of hexosyl- and sialyltransferase domains in the bifunctional capsule polymerases from Neisseria meningitidis W and Y defines a new sialyltransferase family
- PMID: 25342753
- PMCID: PMC4256332
- DOI: 10.1074/jbc.M114.597773
Dissection of hexosyl- and sialyltransferase domains in the bifunctional capsule polymerases from Neisseria meningitidis W and Y defines a new sialyltransferase family
Abstract
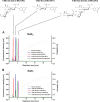
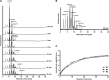
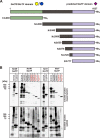
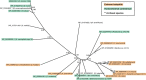
Crucial virulence determinants of disease causing Neisseria meningitidis species are their extracellular polysaccharide capsules. In the serogroups W and Y, these are heteropolymers of the repeating units (→6)-α-d-Gal-(1→4)-α-Neu5Ac-(2→)n in NmW and (→6)-α-d-Glc-(1→4)-α-Neu5Ac-(2→)n in NmY. The capsule polymerases, SiaDW and SiaDY, which synthesize these highly unusual polymers, are composed of two predicted GT-B fold domains separated by a large stretch of amino acids (aa 399-762). We recently showed that residues critical to the hexosyl- and sialyltransferase activity are found in the predicted N-terminal (aa 1-398) and C-terminal (aa 763-1037) GT-B fold domains, respectively. Here we use a mutational approach and synthetic fluorescent substrates to define the boundaries of the hexosyl- and sialyltransferase domains. Our results reveal that the active sialyltransferase domain extends well beyond the predicted C-terminal GT-B domain and defines a new glycosyltransferase family, GT97, in CAZy (Carbohydrate-Active enZYmes Database).
Keywords: Bioinformatics; Capsule Polymerases; Enzyme Catalysis; Fluorescence-based Testing of Glycosyltransferases; Glycosyltransferase; Hexosyltransferases; Neisseria meningitidis Serogroup W and Y; Phylogenetics; Polysaccharide; Sialyltransferases.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Willis L. M., Whitfield C. (2013) Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 378, 35–44 - PubMed
-
- Stephens D. S., Greenwood B., Brandtzaeg P. (2007) Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369, 2196–2210 - PubMed
-
- Whitfield C., Roberts I. S. (1999) Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31, 1307–1319 - PubMed
-
- Whitfield C. (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

