Impact of three commercial feed formulations on farmed gilthead sea bream (Sparus aurata, L.) metabolism as inferred from liver and blood serum proteomics
- PMID: 25342931
- PMCID: PMC4200174
- DOI: 10.1186/s12953-014-0044-3
Impact of three commercial feed formulations on farmed gilthead sea bream (Sparus aurata, L.) metabolism as inferred from liver and blood serum proteomics
Abstract
Background: The zootechnical performance of three different commercial feeds and their impact on liver and serum proteins of gilthead sea bream (Sparus aurata, L.) were assessed in a 12 week feeding trial. The three feeds, named A, B, and C, were subjected to lipid and protein characterization by gas chromatography (GC) and liquid chromatography-tandem mass spectrometry (LC-MS/MS), respectively.
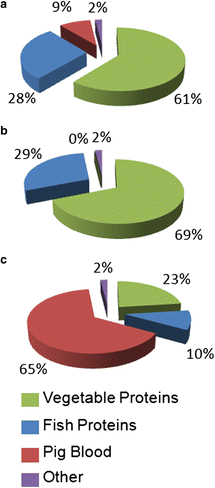
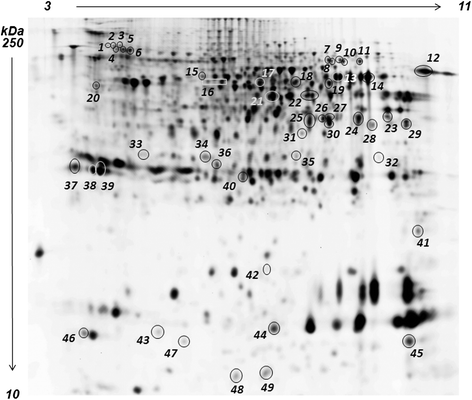
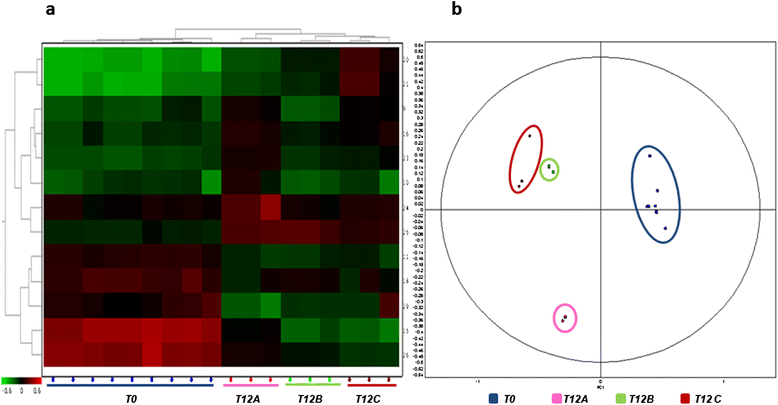
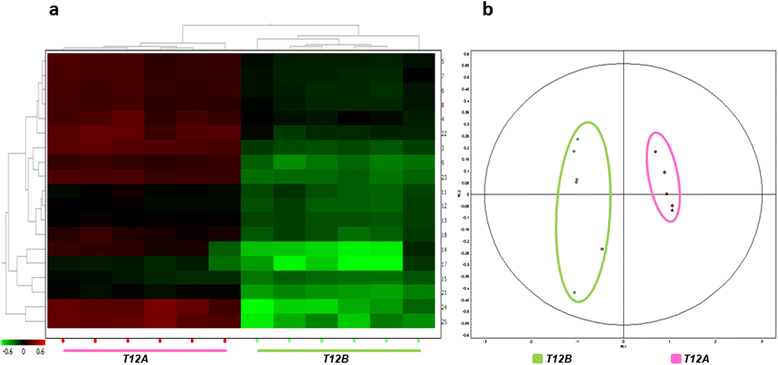
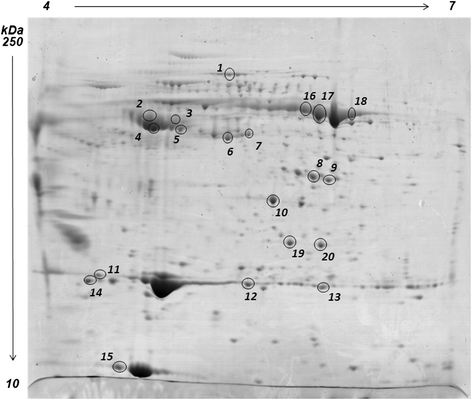
Results: Feed B was higher in fish-derived lipids and proteins, while feeds C and A were higher in vegetable components, although the largest proportion of feed C proteins was represented by pig hemoglobin. According to biometric measurements, the feeds had significantly different impacts on fish growth, producing a higher average weight gain and a lower liver somatic index in feed B over feeds A and C, respectively. 2D DIGE/MS analysis of liver tissue and Ingenuity pathways analysis (IPA) highlighted differential changes in proteins involved in key metabolic pathways of liver, spanning carbohydrate, lipid, protein, and oxidative metabolism. In addition, serum proteomics revealed interesting changes in apolipoproteins, transferrin, warm temperature acclimation-related 65 kDa protein (Wap65), fibrinogen, F-type lectin, and alpha-1-antitrypsin.
Conclusions: This study highlights the contribution of proteomics for understanding and improving the metabolic compatibility of feeds for marine aquaculture, and opens new perspectives for its monitoring with serological tests.
Keywords: 2D DIGE; Aquaculture; Farmed fish; Fish feed; Gilthead sea bream; Ingenuity pathway analysis; Liver proteins; Mass spectrometry; Proteomics; Serum proteins.
Figures


References
-
- FAO . The state of world fisheries and aquaculture. Rome: FAO Fisheries and Aquaculture Department. Food and Agriculture Organization of the United Nations; 2010.
-
- Carter CG, Hauler RC. Fish meal replacement by plant meals in extruded feeds for Atlantic salmon, Salmo salar L. Aquaculture. 2000;185:299–311. doi: 10.1016/S0044-8486(99)00353-1. - DOI
-
- Kaushik SJ, Cravedi JP, Lalles JP, Sumpter J, Fauconneau B, Laroche M. Partial or total replacement of fish meal by soybean protein on growth, protein utilization, potential estrogenic or antigenic effects, cholesterolemia and flesh quality in rainbow trout, Oncorhynchus mykiss. Aquaculture. 1995;133:257–274. doi: 10.1016/0044-8486(94)00403-B. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

