Plant oxidosqualene metabolism: cycloartenol synthase-dependent sterol biosynthesis in Nicotiana benthamiana
- PMID: 25343375
- PMCID: PMC4208727
- DOI: 10.1371/journal.pone.0109156
Plant oxidosqualene metabolism: cycloartenol synthase-dependent sterol biosynthesis in Nicotiana benthamiana
Abstract
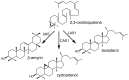
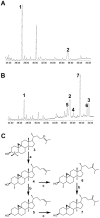
The plant sterol pathway exhibits a major biosynthetic difference as compared with that of metazoans. The committed sterol precursor is the pentacyclic cycloartenol (9β,19-cyclolanost-24-en-3β-ol) and not lanosterol (lanosta-8,24-dien-3β-ol), as it was shown in the late sixties. However, plant genome mining over the last years revealed the general presence of lanosterol synthases encoding sequences (LAS1) in the oxidosqualene cyclase repertoire, in addition to cycloartenol synthases (CAS1) and to non-steroidal triterpene synthases that contribute to the metabolic diversity of C30H50O compounds on earth. Furthermore, plant LAS1 proteins have been unambiguously identified by peptidic signatures and by their capacity to complement the yeast lanosterol synthase deficiency. A dual pathway for the synthesis of sterols through lanosterol and cycloartenol was reported in the model Arabidopsis thaliana, though the contribution of a lanosterol pathway to the production of 24-alkyl-Δ(5)-sterols was quite marginal (Ohyama et al. (2009) PNAS 106, 725). To investigate further the physiological relevance of CAS1 and LAS1 genes in plants, we have silenced their expression in Nicotiana benthamiana. We used virus induced gene silencing (VIGS) based on gene specific sequences from a Nicotiana tabacum CAS1 or derived from the solgenomics initiative (http://solgenomics.net/) to challenge the respective roles of CAS1 and LAS1. In this report, we show a CAS1-specific functional sterol pathway in engineered yeast, and a strict dependence on CAS1 of tobacco sterol biosynthesis.
Conflict of interest statement
Figures



References
-
- Benveniste P, Hirth L, Ourisson G (1966) La biosynthèse des stérols dans les cultures de tissus de tabac cultivés in vitro. II. Particularités de la biosynthèse des phytostérols dans des tissus de tabac cultivés in vitro . Phytochem 5: 45–58.
-
- Hewlins MJE, Ehrhardt JD, Hirth L, Ourisson G (1969) The conversion of 14C-cycloartenol and 14C-lanosterol into phytosterols by cultures of Nicotiana tabacum . Eur J Biochem 8: 184–188. - PubMed
-
- Raederstorff D, Rohmer M (1987) Sterol biosynthesis via cycloartenol and other biochemical features related to photosynthetic phyla in the amoeba Naegleria lovaniensis and Naegleria gruberi . Eur J Biochem 164: 427–34. - PubMed
-
- Benveniste P (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Biol 55: 429–457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

