Molecular insights into the interaction between Plasmodium falciparum apical membrane antigen 1 and an invasion-inhibitory peptide
- PMID: 25343578
- PMCID: PMC4208761
- DOI: 10.1371/journal.pone.0109674
Molecular insights into the interaction between Plasmodium falciparum apical membrane antigen 1 and an invasion-inhibitory peptide
Abstract
Apical membrane antigen 1 (AMA1) of the human malaria parasite Plasmodium falciparum has been implicated in invasion of the host erythrocyte. It interacts with malarial rhoptry neck (RON) proteins in the moving junction that forms between the host cell and the invading parasite. Agents that block this interaction inhibit invasion and may serve as promising leads for anti-malarial drug development. The invasion-inhibitory peptide R1 binds to a hydrophobic cleft on AMA1, which is an attractive target site for small molecules that block parasite invasion. In this work, truncation and mutational analyses show that Phe5-Phe9, Phe12 and Arg15 in R1 are the most important residues for high affinity binding to AMA1. These residues interact with two well-defined binding hot spots on AMA1. Computational solvent mapping reveals that one of these hot spots is suitable for small molecule targeting. We also confirm that R1 in solution binds to AMA1 with 1:1 stoichiometry and adopts a secondary structure consistent with the major form of R1 observed in the crystal structure of the complex. Our results provide a basis for designing high affinity inhibitors of the AMA1-RON2 interaction.
Conflict of interest statement
Figures

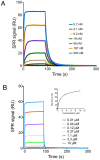
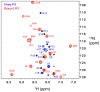
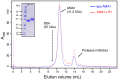

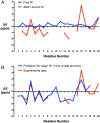
References
-
- World Health Organisation (2013) World Malaria Report 2013.
-
- MacRaild CA, Anders RF, Foley M, Norton RS (2011) Apical membrane antigen 1 as an anti-malarial drug target. Curr Top Med Chem 11: 2039–2047. - PubMed
-
- Miller LH, Baruch DI, Marsh K, Doumbo OK (2002) The pathogenic basis of malaria. Nature 415: 673–679. - PubMed
-
- Tonkin ML, Roques M, Lamarque MH, Pugniere M, Douguet D, et al. (2011) Host cell invasion by apicomplexan parasites: insights from the co-structure of AMA1 with a RON2 peptide. Science 333: 463–467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

