Molecular and metabolic adaptations of Lactococcus lactis at near-zero growth rates
- PMID: 25344239
- PMCID: PMC4272703
- DOI: 10.1128/AEM.02484-14
Molecular and metabolic adaptations of Lactococcus lactis at near-zero growth rates
Abstract
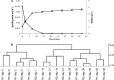
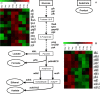
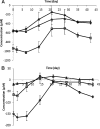
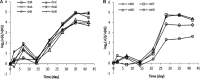
This paper describes the molecular and metabolic adaptations of Lactococcus lactis during the transition from a growing to a near-zero growth state by using carbon-limited retentostat cultivation. Transcriptomic analyses revealed that metabolic patterns shifted between lactic- and mixed-acid fermentations during retentostat cultivation, which appeared to be controlled at the level of transcription of the corresponding pyruvate dissipation-encoding genes. During retentostat cultivation, cells continued to consume several amino acids but also produced specific amino acids, which may derive from the conversion of glycolytic intermediates. We identify a novel motif containing CTGTCAG in the upstream regions of several genes related to amino acid conversion, which we propose to be the target site for CodY in L. lactis KF147. Finally, under extremely low carbon availability, carbon catabolite repression was progressively relieved and alternative catabolic functions were found to be highly expressed, which was confirmed by enhanced initial acidification rates on various sugars in cells obtained from near-zero-growth cultures. The present integrated transcriptome and metabolite (amino acids and previously reported fermentation end products) study provides molecular understanding of the adaptation of L. lactis to conditions supporting low growth rates and expands our earlier analysis of the quantitative physiology of this bacterium at near-zero growth rates toward gene regulation patterns involved in zero-growth adaptation.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Hugas M, Monfort JM. 1997. Bacterial starter cultures for meat fermentation. Food Chem 59:547–554. doi: 10.1016/S0308-8146(97)00005-8. - DOI
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

