Genotyping-by-sequencing based intra-specific genetic map refines a ''QTL-hotspot" region for drought tolerance in chickpea
- PMID: 25344290
- PMCID: PMC4361754
- DOI: 10.1007/s00438-014-0932-3
Genotyping-by-sequencing based intra-specific genetic map refines a ''QTL-hotspot" region for drought tolerance in chickpea
Abstract
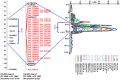
To enhance the marker density in the "QTL-hotspot" region, harboring several QTLs for drought tolerance-related traits identified on linkage group 04 (CaLG04) in chickpea recombinant inbred line (RIL) mapping population ICC 4958 × ICC 1882, a genotyping-by-sequencing approach was adopted. In total, 6.24 Gb data from ICC 4958, 5.65 Gb data from ICC 1882 and 59.03 Gb data from RILs were generated, which identified 828 novel single-nucleotide polymorphisms (SNPs) for genetic mapping. Together with these new markers, a high-density intra-specific genetic map was developed that comprised 1,007 marker loci spanning a distance of 727.29 cM. QTL analysis using the extended genetic map along with precise phenotyping data for 20 traits collected over one to seven seasons identified 49 SNP markers in the "QTL-hotspot" region. These efforts have refined the "QTL-hotspot" region to 14 cM. In total, 164 main-effect QTLs including 24 novel QTLs were identified. In addition, 49 SNPs integrated in the "QTL-hotspot" region were converted into cleaved amplified polymorphic sequence (CAPS) and derived CAPS (dCAPS) markers which can be used in marker-assisted breeding.
Figures



References
-
- Dodig D, Zorić M, Kandić V, Perović D, Šurlan-Momirović G. Comparison of responses to drought stress of 100 wheat accessions and landraces to identify opportunities for improving wheat drought resistance. Plant Breed. 2012;131:369–379. doi: 10.1111/j.1439-0523.2011.01941.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

