The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes
- PMID: 25344531
- PMCID: PMC4234600
- DOI: 10.1073/pnas.1418564111
The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes
Abstract
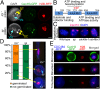
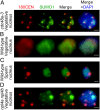
Centromeres mediate chromosome segregation and are defined by the centromere-specific histone H3 variant (CenH3)/centromere protein A (CENP-A). Removal of CenH3 from centromeres is a general property of terminally differentiated cells, and the persistence of CenH3 increases the risk of diseases such as cancer. However, active mechanisms of centromere disassembly are unknown. Nondividing Arabidopsis pollen vegetative cells, which transport engulfed sperm by extended tip growth, undergo loss of CenH3; centromeric heterochromatin decondensation; and bulk activation of silent rRNA genes, accompanied by their translocation into the nucleolus. Here, we show that these processes are blocked by mutations in the evolutionarily conserved AAA-ATPase molecular chaperone, CDC48A, homologous to yeast Cdc48 and human p97 proteins, both of which are implicated in ubiquitin/small ubiquitin-like modifier (SUMO)-targeted protein degradation. We demonstrate that CDC48A physically associates with its heterodimeric cofactor UFD1-NPL4, known to bind ubiquitin and SUMO, as well as with SUMO1-modified CenH3 and mutations in NPL4 phenocopy cdc48a mutations. In WT vegetative cell nuclei, genetically unlinked ribosomal DNA (rDNA) loci are uniquely clustered together within the nucleolus and all major rRNA gene variants, including those rDNA variants silenced in leaves, are transcribed. In cdc48a mutant vegetative cell nuclei, however, these rDNA loci frequently colocalized with condensed centromeric heterochromatin at the external periphery of the nucleolus. Our results indicate that the CDC48A(NPL4) complex actively removes sumoylated CenH3 from centromeres and disrupts centromeric heterochromatin to release bulk rRNA genes into the nucleolus for ribosome production, which fuels single nucleus-driven pollen tube growth and is essential for plant reproduction.
Keywords: centromere disassembly; chromosome dynamics; heterochromatin decondensation; pollen tip growth; rDNA activation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana.PLoS Genet. 2011 Jun;7(6):e1002121. doi: 10.1371/journal.pgen.1002121. Epub 2011 Jun 9. PLoS Genet. 2011. PMID: 21695238 Free PMC article.
-
The Chaperone NASP Contributes to de Novo Deposition of the Centromeric Histone Variant CENH3 in Arabidopsis Early Embryogenesis.Plant Cell Physiol. 2024 Jul 30;65(7):1135-1148. doi: 10.1093/pcp/pcae030. Plant Cell Physiol. 2024. PMID: 38597891 Free PMC article.
-
The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana.Genetics. 2010 Oct;186(2):461-71. doi: 10.1534/genetics.110.120337. Epub 2010 Jul 13. Genetics. 2010. PMID: 20628040 Free PMC article.
-
Heterochromatin dynamics during developmental transitions in Arabidopsis - a focus on ribosomal DNA loci.Gene. 2013 Aug 15;526(1):39-45. doi: 10.1016/j.gene.2013.01.060. Epub 2013 Feb 12. Gene. 2013. PMID: 23410919 Review.
-
Centromeres and kinetochores of Brassicaceae.Chromosome Res. 2014 Jun;22(2):135-52. doi: 10.1007/s10577-014-9422-z. Chromosome Res. 2014. PMID: 24801345 Review.
Cited by
-
Oxidative and salt stresses alter the 26S proteasome holoenzyme and associated protein profiles in Arabidopsis thaliana.BMC Plant Biol. 2021 Oct 25;21(1):486. doi: 10.1186/s12870-021-03234-9. BMC Plant Biol. 2021. PMID: 34696730 Free PMC article.
-
Stable centromere positioning in diverse sequence contexts of complex and satellite centromeres of maize and wild relatives.Genome Biol. 2017 Jun 21;18(1):121. doi: 10.1186/s13059-017-1249-4. Genome Biol. 2017. PMID: 28637491 Free PMC article.
-
A simple and robust protocol for immunostaining Arabidopsis pollen nuclei.Plant Reprod. 2019 Mar;32(1):39-43. doi: 10.1007/s00497-018-00360-7. Epub 2019 Jan 22. Plant Reprod. 2019. PMID: 30671645
-
SUMO-interacting motifs (SIMs) in Polo-like kinase 1-interacting checkpoint helicase (PICH) ensure proper chromosome segregation during mitosis.Cell Cycle. 2016 Aug 17;15(16):2135-2144. doi: 10.1080/15384101.2016.1191713. Epub 2016 May 26. Cell Cycle. 2016. PMID: 27230136 Free PMC article.
-
SUMO control of centromere homeostasis.Front Cell Dev Biol. 2023 Apr 27;11:1193192. doi: 10.3389/fcell.2023.1193192. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37181753 Free PMC article. Review.
References
-
- Hennig W. Heterochromatin. Chromosoma. 1999;108(1):1–9. - PubMed
-
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell. 2002;108(4):489–500. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

