CRISPR-Cas systems in the marine actinomycete Salinispora: linkages with phage defense, microdiversity and biogeography
- PMID: 25344663
- PMCID: PMC4223832
- DOI: 10.1186/1471-2164-15-936
CRISPR-Cas systems in the marine actinomycete Salinispora: linkages with phage defense, microdiversity and biogeography
Abstract
Background: Prokaryotic CRISPR-Cas systems confer resistance to viral infection and thus mediate bacteria-phage interactions. However, the distribution and functional diversity of CRISPRs among environmental bacteria remains largely unknown. Here, comparative genomics of 75 Salinispora strains provided insight into the diversity and distribution of CRISPR-Cas systems in a cosmopolitan marine actinomycete genus.
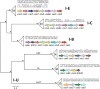
Results: CRISPRs were found in all Salinispora strains, with the majority containing multiple loci and different Cas array subtypes. Of the six subtypes identified, three have not been previously described. A lower prophage frequency in S. arenicola was associated with a higher fraction of spacers matching Salinispora prophages compared to S. tropica, suggesting differing defensive capacities between Salinispora species. The occurrence of related prophages in strains from distant locations, as well as spacers matching those prophages inserted throughout spacer arrays, indicate recurring encounters with widely distributed phages over time. Linkages of CRISPR features with Salinispora microdiversity pointed to subclade-specific contacts with mobile genetic elements (MGEs). This included lineage-specific spacer deletions or insertions, which may reflect weak selective pressures to maintain immunity or distinct temporal interactions with MGEs, respectively. Biogeographic patterns in spacer and prophage distributions support the concept that Salinispora spp. encounter localized MGEs. Moreover, the presence of spacers matching housekeeping genes suggests that CRISPRs may have functions outside of viral defense.
Conclusions: This study provides a comprehensive examination of CRISPR-Cas systems in a broadly distributed group of environmental bacteria. The ubiquity and diversity of CRISPRs in Salinispora suggests that CRISPR-mediated interactions with MGEs represent a major force in the ecology and evolution of this cosmopolitan marine actinomycete genus.
Figures

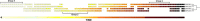

Similar articles
-
Biogeography of the marine actinomycete Salinispora.Environ Microbiol. 2006 Nov;8(11):1881-8. doi: 10.1111/j.1462-2920.2006.01093.x. Environ Microbiol. 2006. PMID: 17014488
-
Atypical organizations and epistatic interactions of CRISPRs and cas clusters in genomes and their mobile genetic elements.Nucleic Acids Res. 2020 Jan 24;48(2):748-760. doi: 10.1093/nar/gkz1091. Nucleic Acids Res. 2020. PMID: 31745554 Free PMC article.
-
Culture-dependent and culture-independent diversity within the obligate marine actinomycete genus Salinispora.Appl Environ Microbiol. 2005 Nov;71(11):7019-28. doi: 10.1128/AEM.71.11.7019-7028.2005. Appl Environ Microbiol. 2005. PMID: 16269737 Free PMC article.
-
Inhibition of CRISPR-Cas systems by mobile genetic elements.Curr Opin Microbiol. 2017 Jun;37:120-127. doi: 10.1016/j.mib.2017.06.003. Epub 2017 Jun 29. Curr Opin Microbiol. 2017. PMID: 28668720 Free PMC article. Review.
-
Meet the Anti-CRISPRs: Widespread Protein Inhibitors of CRISPR-Cas Systems.CRISPR J. 2019 Feb;2(1):23-30. doi: 10.1089/crispr.2018.0052. CRISPR J. 2019. PMID: 31021234 Review.
Cited by
-
Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR-Associated Protein and Its Utility All at Sea: Status, Challenges, and Prospects.Microorganisms. 2024 Jan 6;12(1):118. doi: 10.3390/microorganisms12010118. Microorganisms. 2024. PMID: 38257946 Free PMC article. Review.
-
Pan genome and CRISPR analyses of the bacterial fish pathogen Moritella viscosa.BMC Genomics. 2017 Apr 20;18(1):313. doi: 10.1186/s12864-017-3693-7. BMC Genomics. 2017. PMID: 28427330 Free PMC article.
-
CRISPR-Cas in actinomycetes: still a lot to be discovered.Microlife. 2025 Jun 12;6:uqaf010. doi: 10.1093/femsml/uqaf010. eCollection 2025. Microlife. 2025. PMID: 40626243 Free PMC article. Review.
-
Aquaculture as a source of empirical evidence for coevolution between CRISPR-Cas and phage.Philos Trans R Soc Lond B Biol Sci. 2019 May 13;374(1772):20180100. doi: 10.1098/rstb.2018.0100. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 30905289 Free PMC article. Review.
-
Novel Viral Communities Potentially Assisting in Carbon, Nitrogen, and Sulfur Metabolism in the Upper Slope Sediments of Mariana Trench.mSystems. 2022 Feb 22;7(1):e0135821. doi: 10.1128/msystems.01358-21. Epub 2022 Jan 4. mSystems. 2022. PMID: 35089086 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

