Human-induced evolution caught in action: SNP-array reveals rapid amphi-atlantic spread of pesticide resistance in the salmon ecotoparasite Lepeophtheirus salmonis
- PMID: 25344698
- PMCID: PMC4223847
- DOI: 10.1186/1471-2164-15-937
Human-induced evolution caught in action: SNP-array reveals rapid amphi-atlantic spread of pesticide resistance in the salmon ecotoparasite Lepeophtheirus salmonis
Abstract
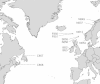
Background: The salmon louse, Lepeophtheirus salmonis, is an ectoparasite of salmonids that causes huge economic losses in salmon farming, and has also been causatively linked with declines of wild salmonid populations. Lice control on farms is reliant upon a few groups of pesticides that have all shown time-limited efficiency due to resistance development. However, to date, this example of human-induced evolution is poorly documented at the population level due to the lack of molecular tools. As such, important evolutionary and management questions, linked to the development and dispersal of pesticide resistance in this parasite, remain unanswered. Here, we introduce the first Single Nucleotide Polymorphism (SNP) array for the salmon louse, which includes 6000 markers, and present a population genomic scan using this array on 576 lice from twelve farms distributed across the North Atlantic.
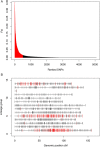
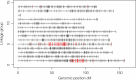
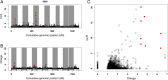
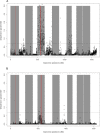
Results: Our results support the hypothesis of a single panmictic population of lice in the Atlantic, and importantly, revealed very strong selective sweeps on linkage groups 1 and 5. These sweeps included candidate genes potentially connected to pesticide resistance. After genotyping a further 576 lice from 12 full sibling families, a genome-wide association analysis established a highly significant association between the major sweep on linkage group 5 and resistance to emamectin benzoate, the most widely used pesticide in salmonid aquaculture for more than a decade.
Conclusions: The analysis of conserved haplotypes across samples from the Atlantic strongly suggests that emamectin benzoate resistance developed at a single source, and rapidly spread across the Atlantic within the period 1999 when the chemical was first introduced, to 2010 when samples for the present study were obtained. These results provide unique insights into the development and spread of pesticide resistance in the marine environment, and identify a small genomic region strongly linked to emamectin benzoate resistance. Finally, these results have highly significant implications for the way pesticide resistance is considered and managed within the aquaculture industry.
Figures


References
-
- Bjørn PA, Finstad B. The development of salmon lice (Lepeophtheirus salmonis) on artificially infected post smolts of sea trout (Salmo trutta) Can J Zool. 1998;76:970–977. doi: 10.1139/cjz-76-5-970. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

