Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress
- PMID: 25344756
- PMCID: PMC5224936
- DOI: 10.1038/ncb3054
Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress
Abstract
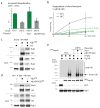
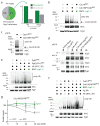
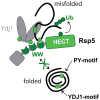
The heat-shock response is a complex cellular program that induces major changes in protein translation, folding and degradation to alleviate toxicity caused by protein misfolding. Although heat shock has been widely used to study proteostasis, it remained unclear how misfolded proteins are targeted for proteolysis in these conditions. We found that Rsp5 and its mammalian homologue Nedd4 are important E3 ligases responsible for the increased ubiquitylation induced by heat stress. We determined that Rsp5 ubiquitylates mainly cytosolic misfolded proteins upon heat shock for proteasome degradation. We found that ubiquitylation of heat-induced substrates requires the Hsp40 co-chaperone Ydj1 that is further associated with Rsp5 upon heat shock. In addition, ubiquitylation is also promoted by PY Rsp5-binding motifs found primarily in the structured regions of stress-induced substrates, which can act as heat-induced degrons. Our results support a bipartite recognition mechanism combining direct and chaperone-dependent ubiquitylation of misfolded cytosolic proteins by Rsp5.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
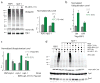
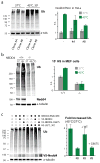

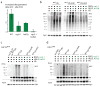
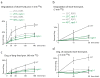
Comment in
-
Rsp5/Nedd4 clears cells of heat-damaged proteins.Nat Cell Biol. 2014 Dec;16(12):1130-2. doi: 10.1038/ncb3079. Nat Cell Biol. 2014. PMID: 25434464
References
-
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. - PubMed
-
- Comyn SA, Chan GT, Mayor T. False start: Cotranslational protein ubiquitination and cytosolic protein quality control. J Proteomics. 2013 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

