The orthologous Tbx transcription factors Omb and TBX2 induce epithelial cell migration and extrusion in vivo without involvement of matrix metalloproteinases
- PMID: 25344916
- PMCID: PMC4322970
- DOI: 10.18632/oncotarget.2426
The orthologous Tbx transcription factors Omb and TBX2 induce epithelial cell migration and extrusion in vivo without involvement of matrix metalloproteinases
Abstract
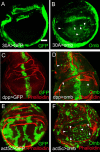
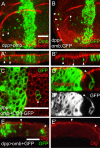
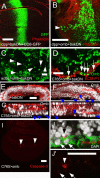
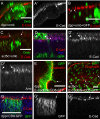
The transcription factors TBX2 and TBX3 are overexpressed in various human cancers. Here, we investigated the effect of overexpressing the orthologous Tbx genes Drosophila optomotor-blind (omb) and human TBX2 in the epithelium of the Drosophila wing imaginal disc and observed two types of cell motility. Omb/TBX2 overexpressing cells could move within the plane of the epithelium. Invasive cells migrated long-distance as single cells retaining or regaining normal cell shape and apico-basal polarity in spite of attenuated apical DE-cadherin concentration. Inappropriate levels of DE-cadherin were sufficient to drive cell migration in the wing disc epithelium. Omb/TBX2 overexpression and reduced DE-cadherin-dependent adhesion caused the formation of actin-rich lateral cell protrusions. Omb/TBX2 overexpressing cells could also delaminate basally, penetrating the basal lamina, however, without degradation of extracellular matrix. Expression of Timp, an inhibitor of matrix metalloproteases, blocked neither intraepithelial motility nor basal extrusion. Our results reveal an MMP-independent mechanism of cell invasion and suggest a conserved role of Tbx2-related proteins in cell invasion and metastasis-related processes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

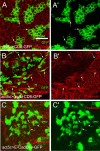
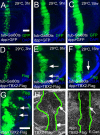

References
-
- Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, Wu T, Niinobe M, Yoshikawa K, Hannigan GE, Halaban R. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64(15):5270–5282. - PubMed
-
- Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 2005;65(6):2260–2268. - PubMed
-
- Boyd SC, Mijatov B, Pupo GM, Tran SL, Gowrishankar K, Shaw HM, Goding CR, Scolyer RA, Mann GJ, Kefford RF, Rizos H, Becker TM. Oncogenic B-RAF(V600E) Signaling Induces the T-Box3 Transcriptional Repressor to Repress E-Cadherin and Enhance Melanoma Cell Invasion. J Invest Dermatol. 2013;133(5):1269–1277. - PMC - PubMed
-
- Abrahams A, Parker MI, Prince S. The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life. 2010;62(2):92–102. - PubMed
-
- Rowley M, Grothey E, Couch FJ. The role of Tbx2 and Tbx3 in mammary development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2004;9(2):109–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

