Tryptophan-catabolizing enzymes - party of three
- PMID: 25346733
- PMCID: PMC4191572
- DOI: 10.3389/fimmu.2014.00485
Tryptophan-catabolizing enzymes - party of three
Abstract
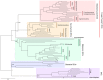
Indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) are tryptophan-degrading enzymes that have independently evolved to catalyze the first step in tryptophan catabolism via the kynurenine pathway (KP). The depletion of tryptophan and formation of KP metabolites modulates the activity of the mammalian immune, reproductive, and central nervous systems. IDO and TDO enzymes can have overlapping or distinct functions depending on their expression patterns. The expression of TDO and IDO enzymes in mammals differs not only by tissue/cellular localization but also by their induction by distinct stimuli. To add to the complexity, these genes also have undergone duplications in some organisms leading to multiple isoforms of IDO or TDO. For example, many vertebrates, including all mammals, have acquired two IDO genes via gene duplication, although the IDO1-like gene has been lost in some lower vertebrate lineages. Gene duplications can allow the homologs to diverge and acquire different properties to the original gene. There is evidence for IDO enzymes having differing enzymatic characteristics, signaling properties, and biological functions. This review analyzes the evolutionary convergence of IDO and TDO enzymes as tryptophan-catabolizing enzymes and the divergent evolution of IDO homologs to generate an enzyme family with diverse characteristics not possessed by TDO enzymes, with an emphasis on the immune system.
Keywords: convergent evolution; divergent evolution; gene duplication; immunoregulation; indoleamine 2,3-dioxygenase; tryptophan 2,3-dioxygenase.
Figures



References
-
- Leklem JE. Quantitative aspects of tryptophan metabolism in humans and other species: a review. Am J Clin Nutr (1971) 24:659–72 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

