A dehydrin-dehydrin interaction: the case of SK3 from Opuntia streptacantha
- PMID: 25346739
- PMCID: PMC4193212
- DOI: 10.3389/fpls.2014.00520
A dehydrin-dehydrin interaction: the case of SK3 from Opuntia streptacantha
Abstract
Dehydrins belongs to a large group of highly hydrophilic proteins known as Late Embryogenesis Abundant (LEA) proteins. It is well known that dehydrins are intrinsically disordered plant proteins that accumulate during the late stages of embryogenesis and in response to abiotic stresses; however, the molecular mechanisms by which their functions are carried out are still unclear. We have previously reported that transgenic Arabidopsis plants overexpressing an Opuntia streptacantha SK3 dehydrin (OpsDHN1) show enhanced tolerance to freezing stress. Herein, we show using a split-ubiquitin yeast two-hybrid system that OpsDHN1 dimerizes. We found that the deletion of regions containing K-segments and the histidine-rich region in the OpsDHN1 protein affects dimer formation. Not surprisingly, in silico protein sequence analysis suggests that OpsDHN1 is an intrinsically disordered protein, an observation that was confirmed by circular dichroism and gel filtration of the recombinantly expressed protein. The addition of zinc triggered the association of recombinantly expressed OpsDHN1 protein, likely through its histidine-rich motif. These data brings new insights about the molecular mechanism of the OpsDHN1 SK3-dehydrin.
Keywords: K-segments; SK3-dehydrin; histidine-rich region; homodimer; intrinsically disordered proteins; yeast two-hybrid.
Figures
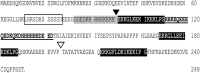




References
-
- Close T. J. (1996). Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Planta 97, 795–803 10.1111/j.1399-3054.1996.tb00546.x - DOI
-
- Close T. J. (1997). Dehydrins: a commonalty in the response of plants to dehydration and low temperature. Physiol. Planta 100, 291–296 10.1111/j.1399-3054.1997.tb04785.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

