Isolation of epithelial, endothelial, and immune cells from lungs of transgenic mice with oncogene-induced lung adenocarcinomas
- PMID: 25347711
- PMCID: PMC4491125
- DOI: 10.1165/rcmb.2014-0312MA
Isolation of epithelial, endothelial, and immune cells from lungs of transgenic mice with oncogene-induced lung adenocarcinomas
Abstract
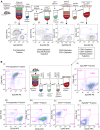
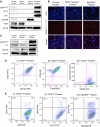
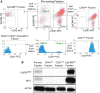
Genetically engineered mouse models of lung adenocarcinoma have proven invaluable for understanding mechanisms of tumorigenesis, therapy response, and drug resistance. However, mechanistic studies focused on studying these processes in tumor-bearing mouse lungs are confounded by the fact that, in most cases, relevant signaling pathways are analyzed in whole-lung preparations, which are composed of a heterogeneous mixture of cells. Given our increasing knowledge about the roles played by different subpopulations of cells in the development of lung adenocarcinoma, separating the major cellular compartments of the tumor microenvironment is recommended to allow for a precise analysis of relevant pathways in each isolated cell type. In this study, we optimized magnetic- and fluorescence-based isolation protocols to segregate lung epithelial (CD326/epithelial cell adhesion molecule-positive), endothelial (CD31-positive), and immune (CD45-positive) cells, with high purity, from the lungs of transgenic mice with mutant epidermal growth factor receptor-induced lung adenocarcinomas. This approach, which can potentially be extended to additional lung adenocarcinoma models, enables delineation of the molecular features of individual cell types that can be used to gain insight into their roles in lung adenocarcinoma initiation, progression, and response to therapy.
Keywords: epithelial cell adhesion molecule; epithelial cell isolation; lung adenocarcinoma; transgenic mouse models.
Figures
References
-
- Broeckaert F, Bernard A. Clara cell secretory protein (CC16): characteristics and perspectives as lung peripheral biomarker. Clin Exp Allergy. 2000;30:469–475. - PubMed
-
- Ward HE, Nicholas TE. Alveolar type I and type II cells. Aust N Z J Med. 1984;14:731–734. - PubMed
-
- Phelps DS, Floros J. Localization of pulmonary surfactant proteins using immunohistochemistry and tissue in situ hybridization. Exp Lung Res. 1991;17:985–995. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

