The impact of osteoblastic differentiation on osteosarcomagenesis in the mouse
- PMID: 25347737
- PMCID: PMC4411188
- DOI: 10.1038/onc.2014.354
The impact of osteoblastic differentiation on osteosarcomagenesis in the mouse
Abstract
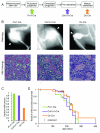
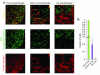
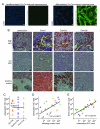
Osteosarcomas remain an enigmatic group of malignancies that share in common the presence of transformed cells producing osteoid matrix, even if these cells comprise a minority of the tumor volume. The differentiation state of osteosarcomas has therefore become a topic of interest and challenge to those who study this disease. In order to test how the cell of origin contributes to the final state of differentiation in the transformed cells, we compared the relative tumorigenicity of Cre-LoxP conditional disruption of the cell cycle checkpoint tumor-suppressor genes Trp53 and Rb1 using Prx1-Cre, Collagen-1α1-Cre and Osteocalcin-Cre to transform undifferentiated mesenchyme, preosteoblasts and mature osteoblasts, respectively. The Prx1 and Col1α1 lineages developed tumors with nearly complete penetrance, as anticipated. Osteosarcomas also developed in 44% of Oc-Cre;Rb1(fl/fl);Trp53(fl/fl) mice. We confirmed using 5-ethynyl-2'-deoxyuridine click chemistry that the Oc-Cre lineage includes very few actively cycling cells. By assessing radiographic mineralization and histological osteoid production, the differentiation state of tumors did not correlate with the differentiation state of the lineage of origin. Some of the osteocalcin-lineage-derived osteosarcomas were among the least osteoblastic. Osteocalcin immunohistochemistry in tumors correlated well with the expression of DNA methyl transferases, suggesting that silencing of these epigenetic regulators may influence the final differentiation state of an osteosarcoma. Transformation of differentiated, minimally proliferative osteoblasts is possible but may require such an epigenetic reprogramming that the tumors no longer resemble their differentiated origins.
Figures

References
-
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11. - PubMed
-
- Sharkey FE. Morphometric analysis of differentiation in human breast carcinoma. Tumor heterogeneity. Archives of pathology & laboratory medicine. 1983;107(8):411–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

