MitProNet: A knowledgebase and analysis platform of proteome, interactome and diseases for mammalian mitochondria
- PMID: 25347823
- PMCID: PMC4210245
- DOI: 10.1371/journal.pone.0111187
MitProNet: A knowledgebase and analysis platform of proteome, interactome and diseases for mammalian mitochondria
Abstract
Mitochondrion plays a central role in diverse biological processes in most eukaryotes, and its dysfunctions are critically involved in a large number of diseases and the aging process. A systematic identification of mitochondrial proteomes and characterization of functional linkages among mitochondrial proteins are fundamental in understanding the mechanisms underlying biological functions and human diseases associated with mitochondria. Here we present a database MitProNet which provides a comprehensive knowledgebase for mitochondrial proteome, interactome and human diseases. First an inventory of mammalian mitochondrial proteins was compiled by widely collecting proteomic datasets, and the proteins were classified by machine learning to achieve a high-confidence list of mitochondrial proteins. The current version of MitProNet covers 1124 high-confidence proteins, and the remainders were further classified as middle- or low-confidence. An organelle-specific network of functional linkages among mitochondrial proteins was then generated by integrating genomic features encoded by a wide range of datasets including genomic context, gene expression profiles, protein-protein interactions, functional similarity and metabolic pathways. The functional-linkage network should be a valuable resource for the study of biological functions of mitochondrial proteins and human mitochondrial diseases. Furthermore, we utilized the network to predict candidate genes for mitochondrial diseases using prioritization algorithms. All proteins, functional linkages and disease candidate genes in MitProNet were annotated according to the information collected from their original sources including GO, GEO, OMIM, KEGG, MIPS, HPRD and so on. MitProNet features a user-friendly graphic visualization interface to present functional analysis of linkage networks. As an up-to-date database and analysis platform, MitProNet should be particularly helpful in comprehensive studies of complicated biological mechanisms underlying mitochondrial functions and human mitochondrial diseases. MitProNet is freely accessible at http://bio.scu.edu.cn:8085/MitProNet.
Conflict of interest statement
Figures
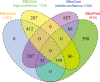
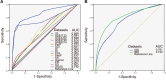





Similar articles
-
InterMitoBase: an annotated database and analysis platform of protein-protein interactions for human mitochondria.BMC Genomics. 2011 Jun 30;12:335. doi: 10.1186/1471-2164-12-335. BMC Genomics. 2011. PMID: 21718467 Free PMC article.
-
The draft nuclear genome sequence and predicted mitochondrial proteome of Andalucia godoyi, a protist with the most gene-rich and bacteria-like mitochondrial genome.BMC Biol. 2020 Mar 2;18(1):22. doi: 10.1186/s12915-020-0741-6. BMC Biol. 2020. PMID: 32122349 Free PMC article.
-
PCHM: A bioinformatic resource for high-throughput human mitochondrial proteome searching and comparison.Comput Biol Med. 2009 Aug;39(8):689-96. doi: 10.1016/j.compbiomed.2009.05.006. Epub 2009 Jun 21. Comput Biol Med. 2009. PMID: 19541297
-
Mitochondrial proteome research: the road ahead.Nat Rev Mol Cell Biol. 2024 Jan;25(1):65-82. doi: 10.1038/s41580-023-00650-7. Epub 2023 Sep 29. Nat Rev Mol Cell Biol. 2024. PMID: 37773518 Free PMC article. Review.
-
Exploring mitochondrial system properties of neurodegenerative diseases through interactome mapping.J Proteomics. 2014 Apr 4;100:8-24. doi: 10.1016/j.jprot.2013.11.008. Epub 2013 Nov 18. J Proteomics. 2014. PMID: 24262152 Review.
Cited by
-
Magnesium induces preconditioning of the neonatal brain via profound mitochondrial protection.J Cereb Blood Flow Metab. 2019 Jun;39(6):1038-1055. doi: 10.1177/0271678X17746132. Epub 2017 Dec 5. J Cereb Blood Flow Metab. 2019. PMID: 29206066 Free PMC article.
-
Mitochondrial Changes in β0-Thalassemia/Hb E Disease.PLoS One. 2016 Apr 19;11(4):e0153831. doi: 10.1371/journal.pone.0153831. eCollection 2016. PLoS One. 2016. PMID: 27092778 Free PMC article.
References
-
- Chan DC (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125: 1241–1252. - PubMed
-
- Traish AM, Abdallah B, Yu G (2011) Androgen deficiency and mitochondrial dysfunction: implications for fatigue, muscle dysfunction, insulin resistance, diabetes, and cardiovascular disease. Hormone Molecular Biology and Clinical Investigation 8: 431–444. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

