Genome3D: exploiting structure to help users understand their sequences
- PMID: 25348407
- PMCID: PMC4384030
- DOI: 10.1093/nar/gku973
Genome3D: exploiting structure to help users understand their sequences
Abstract
Genome3D (http://www.genome3d.eu) is a collaborative resource that provides predicted domain annotations and structural models for key sequences. Since introducing Genome3D in a previous NAR paper, we have substantially extended and improved the resource. We have annotated representatives from Pfam families to improve coverage of diverse sequences and added a fast sequence search to the website to allow users to find Genome3D-annotated sequences similar to their own. We have improved and extended the Genome3D data, enlarging the source data set from three model organisms to 10, and adding VIVACE, a resource new to Genome3D. We have analysed and updated Genome3D's SCOP/CATH mapping. Finally, we have improved the superposition tools, which now give users a more powerful interface for investigating similarities and differences between structural models.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
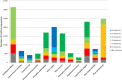
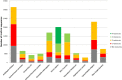

References
-
- Lewis T.E., Sillitoe I., Andreeva A., Blundell T.L., Buchan D.W., Chothia C., Cuff A., Dana J.M., Filippis I., Gough J., et al. Genome3D: a UK collaborative project to annotate genomic sequences with predicted 3D structures based on SCOP and CATH domains. Nucleic Acids Res. 2013;41:D499–D507. - PMC - PubMed
-
- Shi J., Blundell T.L., Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. - PubMed
-
- Lobley A., Sadowski M.I., Jones D.T. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25:1761–1767. - PubMed
Publication types
MeSH terms
Grants and funding
- MC_U105192716/MRC_/Medical Research Council/United Kingdom
- BB/F020481/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/G022771/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I025050/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

