CATH: comprehensive structural and functional annotations for genome sequences
- PMID: 25348408
- PMCID: PMC4384018
- DOI: 10.1093/nar/gku947
CATH: comprehensive structural and functional annotations for genome sequences
Abstract
The latest version of the CATH-Gene3D protein structure classification database (4.0, http://www.cathdb.info) provides annotations for over 235,000 protein domain structures and includes 25 million domain predictions. This article provides an update on the major developments in the 2 years since the last publication in this journal including: significant improvements to the predictive power of our functional families (FunFams); the release of our 'current' putative domain assignments (CATH-B); a new, strictly non-redundant data set of CATH domains suitable for homology benchmarking experiments (CATH-40) and a number of improvements to the web pages.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


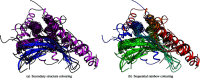

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

