A novel role for protein kinase Kin2 in regulating HAC1 mRNA translocation, splicing, and translation
- PMID: 25348718
- PMCID: PMC4295377
- DOI: 10.1128/MCB.00981-14
A novel role for protein kinase Kin2 in regulating HAC1 mRNA translocation, splicing, and translation
Abstract
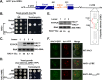
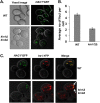
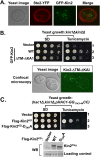
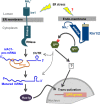
A signaling network called the unfolded protein response (UPR) resolves the protein-folding defects in the endoplasmic reticulum (ER) from yeasts to humans. In the yeast Saccharomyces cerevisiae, the UPR activation involves (i) aggregation of the ER-resident kinase/RNase Ire1 to form an Ire1 focus, (ii) targeting HAC1 pre-mRNA toward the Ire1 focus that cleaves out an inhibitory intron from the mRNA, and (iii) translation of Hac1 protein from the spliced mRNA. Targeting HAC1 mRNA to the Ire1 focus requires a cis-acting bipartite element (3'BE) located at the 3' untranslated leader. Here, we report that the 3'BE plays an additional role in promoting translation from the spliced mRNA. We also report that a high dose of either of two paralogue kinases, Kin1 and Kin2, overcomes the defective UPR caused by a mutation in the 3'BE. These results define a novel role for Kin kinases in the UPR beyond their role in cell polarity and exocytosis. Consistently, targeting, splicing, and translation of HAC1 mRNA are substantially reduced in the kin1Δ kin2Δ strain. Furthermore, we show that Kin2 kinase domain itself is sufficient to activate the UPR, suggesting that Kin2 initiates a signaling cascade to ensure an optimum UPR.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae.Int J Mol Sci. 2019 Jun 12;20(12):2860. doi: 10.3390/ijms20122860. Int J Mol Sci. 2019. PMID: 31212749 Free PMC article. Review.
-
Phosphorylation of Pal2 by the protein kinases Kin1 and Kin2 modulates HAC1 mRNA splicing in the unfolded protein response in yeast.Sci Signal. 2021 May 25;14(684):eaaz4401. doi: 10.1126/scisignal.aaz4401. Sci Signal. 2021. PMID: 34035143 Free PMC article.
-
Messenger RNA targeting to endoplasmic reticulum stress signalling sites.Nature. 2009 Feb 5;457(7230):736-40. doi: 10.1038/nature07641. Epub 2008 Dec 14. Nature. 2009. PMID: 19079237 Free PMC article.
-
Ribosome depurination by ricin leads to inhibition of endoplasmic reticulum stress-induced HAC1 mRNA splicing on the ribosome.J Biol Chem. 2019 Nov 22;294(47):17848-17862. doi: 10.1074/jbc.RA119.009128. Epub 2019 Oct 17. J Biol Chem. 2019. PMID: 31624149 Free PMC article.
-
[The mRNA localization for cytoplasmic splicing].Tanpakushitsu Kakusan Koso. 2009 Dec;54(16 Suppl):2177-83. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089637 Review. Japanese. No abstract available.
Cited by
-
Adaptation to Endoplasmic Reticulum Stress Requires Transphosphorylation within the Activation Loop of Protein Kinases Kin1 and Kin2, Orthologs of Human Microtubule Affinity-Regulating Kinase.Mol Cell Biol. 2018 Nov 13;38(23):e00266-18. doi: 10.1128/MCB.00266-18. Print 2018 Dec 1. Mol Cell Biol. 2018. PMID: 30201804 Free PMC article.
-
Vps34 and TOR Kinases Coordinate HAC1 mRNA Translation in the Presence or Absence of Ire1-Dependent Splicing.Mol Cell Biol. 2021 Jun 23;41(7):e0066220. doi: 10.1128/MCB.00662-20. Epub 2021 Jun 23. Mol Cell Biol. 2021. PMID: 33972394 Free PMC article.
-
Tunicamycin Sensitivity-Suppression by High Gene Dosage Reveals New Functions of the Yeast Hog1 MAP Kinase.Cells. 2019 Jul 12;8(7):710. doi: 10.3390/cells8070710. Cells. 2019. PMID: 31336877 Free PMC article.
-
Substrate priming enhances phosphorylation by the budding yeast kinases Kin1 and Kin2.J Biol Chem. 2018 Nov 23;293(47):18353-18364. doi: 10.1074/jbc.RA118.005651. Epub 2018 Oct 10. J Biol Chem. 2018. PMID: 30305396 Free PMC article.
-
Identification of stress responsive genes by studying specific relationships between mRNA and protein abundance.Heliyon. 2018 Mar 8;4(3):e00558. doi: 10.1016/j.heliyon.2018.e00558. eCollection 2018 Mar. Heliyon. 2018. PMID: 29560469 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
