Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development
- PMID: 25348719
- PMCID: PMC4295379
- DOI: 10.1128/MCB.01054-14
Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development
Abstract
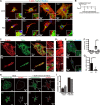
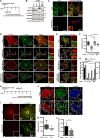
Mitochondria are dynamic organelles, and their fusion and fission regulate cellular signaling, development, and mitochondrial homeostasis, including mitochondrial DNA (mtDNA) distribution. Cardiac myocytes have a specialized cytoplasmic structure where large mitochondria are aligned into tightly packed myofibril bundles; however, recent studies have revealed that mitochondrial dynamics also plays an important role in the formation and maintenance of cardiomyocytes. Here, we precisely analyzed the role of mitochondrial fission in vivo. The mitochondrial fission GTPase, Drp1, is highly expressed in the developing neonatal heart, and muscle-specific Drp1 knockout (Drp1-KO) mice showed neonatal lethality due to dilated cardiomyopathy. The Drp1 ablation in heart and primary cultured cardiomyocytes resulted in severe mtDNA nucleoid clustering and led to mosaic deficiency of mitochondrial respiration. The functional and structural alteration of mitochondria also led to immature myofibril assembly and defective cardiomyocyte hypertrophy. Thus, the dynamics of mtDNA nucleoids regulated by mitochondrial fission is required for neonatal cardiomyocyte development by promoting homogeneous distribution of active mitochondria throughout the cardiomyocytes.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
