Enantioselective regulation of lactate racemization by LarR in Lactobacillus plantarum
- PMID: 25349156
- PMCID: PMC4288682
- DOI: 10.1128/JB.02192-14
Enantioselective regulation of lactate racemization by LarR in Lactobacillus plantarum
Abstract
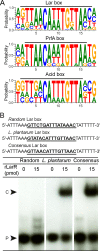
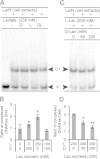
Lactobacillus plantarum is a lactic acid bacterium that produces a racemic mixture of l- and d-lactate from sugar fermentation. The interconversion of lactate isomers is performed by a lactate racemase (Lar) that is transcriptionally controlled by the l-/d-lactate ratio and maximally induced in the presence of l-lactate. We previously reported that the Lar activity depends on the expression of two divergently oriented operons: (i) the larABCDE operon encodes the nickel-dependent lactate racemase (LarA), its maturases (LarBCE), and a lactic acid channel (LarD), and (ii) the larR(MN)QO operon encodes a transcriptional regulator (LarR) and a four-component ABC-type nickel transporter [Lar(MN), in which the M and N components are fused, LarQ, and LarO]. LarR is a novel regulator of the Crp-Fnr family (PrfA group). Here, the role of LarR was further characterized in vivo and in vitro. We show that LarR is a positive regulator that is absolutely required for the expression of Lar activity. Using gel retardation experiments, we demonstrate that LarR binds to a 16-bp palindromic sequence (Lar box motif) that is present in the larR-larA intergenic region. Mutations in the Lar box strongly affect LarR binding and completely abolish transcription from the larA promoter (PlarA). Two half-Lar boxes located between the Lar box and the -35 box of PlarA promote LarR multimerization on DNA, and point mutations within one or both half-Lar boxes inhibit PlarA induction by l-lactate. Gel retardation and footprinting experiments indicate that l-lactate has a positive effect on the binding and multimerization of LarR, while d-lactate antagonizes the positive effect of l-lactate. A possible mechanism of LarR regulation by lactate enantiomers is proposed.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




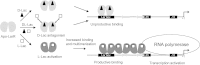
References
-
- Vega Y, Dickneite C, Ripio MT, Bockmann R, Gonzalez-Zorn B, Novella S, Dominguez-Bernal G, Goebel W, Vazquez-Boland JA. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J Bacteriol 180:6655–6660. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

