SCCRO3 (DCUN1D3) antagonizes the neddylation and oncogenic activity of SCCRO (DCUN1D1)
- PMID: 25349211
- PMCID: PMC4263876
- DOI: 10.1074/jbc.M114.585505
SCCRO3 (DCUN1D3) antagonizes the neddylation and oncogenic activity of SCCRO (DCUN1D1)
Abstract
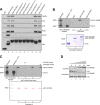
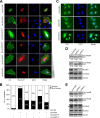
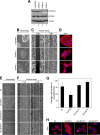
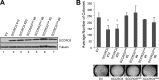
The activity of cullin-RING type ubiquitination E3 ligases is regulated by neddylation, a process analogous to ubiquitination that culminates in covalent attachment of the ubiquitin-like protein Nedd8 to cullins. As a component of the E3 for neddylation, SCCRO/DCUN1D1 plays a key regulatory role in neddylation and, consequently, cullin-RING ligase activity. The essential contribution of SCCRO to neddylation is to promote nuclear translocation of the cullin-ROC1 complex. The presence of a myristoyl sequence in SCCRO3, one of four SCCRO paralogues present in humans that localizes to the membrane, raises questions about its function in neddylation. We found that although SCCRO3 binds to CAND1, cullins, and ROC1, it does not efficiently bind to Ubc12, promote cullin neddylation, or conform to the reaction processivity paradigms, suggesting that SCCRO3 does not have E3 activity. Expression of SCCRO3 inhibits SCCRO-promoted neddylation by sequestering cullins to the membrane, thereby blocking its nuclear translocation. Moreover, SCCRO3 inhibits SCCRO transforming activity. The inhibitory effects of SCCRO3 on SCCRO-promoted neddylation and transformation require both an intact myristoyl sequence and PONY domain, confirming that membrane localization and binding to cullins are required for in vivo functions. Taken together, our findings suggest that SCCRO3 functions as a tumor suppressor by antagonizing the neddylation activity of SCCRO.
Keywords: Head and Neck Cancer; Lung Cancer; Oncogene; Tumor Suppressor Gene; Ubiquitin.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation.J Biol Chem. 2008 Nov 28;283(48):33211-20. doi: 10.1074/jbc.M804440200. Epub 2008 Sep 30. J Biol Chem. 2008. PMID: 18826954 Free PMC article.
-
SCCRO (DCUN1D1) promotes nuclear translocation and assembly of the neddylation E3 complex.J Biol Chem. 2011 Mar 25;286(12):10297-304. doi: 10.1074/jbc.M110.203729. Epub 2011 Jan 19. J Biol Chem. 2011. PMID: 21247897 Free PMC article.
-
The ubiquitin-associated (UBA) domain of SCCRO/DCUN1D1 protein serves as a feedback regulator of biochemical and oncogenic activity.J Biol Chem. 2015 Jan 2;290(1):296-309. doi: 10.1074/jbc.M114.560169. Epub 2014 Nov 19. J Biol Chem. 2015. PMID: 25411243 Free PMC article.
-
Diverse and pivotal roles of neddylation in metabolism and immunity.FEBS J. 2021 Jul;288(13):3884-3912. doi: 10.1111/febs.15584. Epub 2020 Oct 20. FEBS J. 2021. PMID: 33025631 Review.
-
Protein neddylation: beyond cullin-RING ligases.Nat Rev Mol Cell Biol. 2015 Jan;16(1):30-44. doi: 10.1038/nrm3919. Nat Rev Mol Cell Biol. 2015. PMID: 25531226 Free PMC article. Review.
Cited by
-
Anticancer drug discovery by targeting cullin neddylation.Acta Pharm Sin B. 2020 May;10(5):746-765. doi: 10.1016/j.apsb.2019.09.005. Epub 2019 Sep 20. Acta Pharm Sin B. 2020. PMID: 32528826 Free PMC article. Review.
-
Neddylation and deneddylation in cardiac biology.Am J Cardiovasc Dis. 2014 Dec 29;4(4):140-58. eCollection 2014. Am J Cardiovasc Dis. 2014. PMID: 25628956 Free PMC article. Review.
-
The fusion landscape of hepatocellular carcinoma.Mol Oncol. 2019 May;13(5):1214-1225. doi: 10.1002/1878-0261.12479. Epub 2019 Apr 11. Mol Oncol. 2019. PMID: 30903738 Free PMC article.
-
Dcun1d3 is dispensable for spermatogenesis and male fertility in mice.Am J Clin Exp Immunol. 2025 Jun 15;14(3):127-137. doi: 10.62347/BZPE6333. eCollection 2025. Am J Clin Exp Immunol. 2025. PMID: 40689315 Free PMC article.
-
Mild sleep restriction increases endothelial oxidative stress in female persons.Sci Rep. 2023 Sep 16;13(1):15360. doi: 10.1038/s41598-023-42758-y. Sci Rep. 2023. PMID: 37717072 Free PMC article.
References
-
- Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Komander D. (2009) The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37, 937–953 - PubMed
-
- Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 - PubMed
-
- Scheffner M., Nuber U., Huibregtse J. M. (1995) Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373, 81–83 - PubMed
-
- Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

