Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription
- PMID: 25349213
- PMCID: PMC4263867
- DOI: 10.1074/jbc.M114.569244
Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription
Abstract
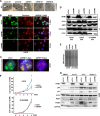
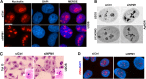
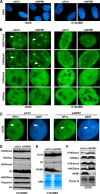
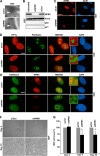
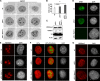
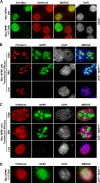
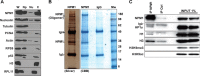
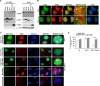
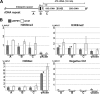
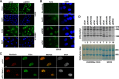
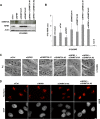
Nucleoli are prominent nuclear structures assembled and organized around actively transcribed ribosomal DNA (rDNA). The nucleolus has emerged as a platform for the organization of chromatin enriched for repressive histone modifications associated with repetitive DNA. NPM1 is a nucleolar protein required for the maintenance of genome stability. However, the role of NPM1 in nucleolar chromatin dynamics and ribosome biogenesis remains unclear. We found that normal fibroblasts and cancer cells depleted of NPM1 displayed deformed nucleoli and a striking rearrangement of perinucleolar heterochromatin, as identified by immunofluorescence staining of trimethylated H3K9, trimethylated H3K27, and heterochromatin protein 1γ (HP1γ/CBX3). By co-immunoprecipitation we found NPM1 associated with HP1γ and core and linker histones. Moreover, NPM1 was required for efficient tethering of HP1γ-enriched chromatin to the nucleolus. We next tested whether the alterations in perinucleolar heterochromatin architecture correlated with a difference in the regulation of rDNA. U1242MG glioma cells depleted of NPM1 presented with altered silver staining of nucleolar organizer regions, coupled to a modest decrease in H3K9 di- and trimethylation at the rDNA promoter. rDNA transcription and cell proliferation were sustained in these cells, indicating that altered organization of heterochromatin was not secondary to inhibition of rDNA transcription. Furthermore, knockdown of DNA methyltransferase DNMT3A markedly enhanced rDNA transcription in NPM1-depleted U1242MG cells. In summary, this study highlights a function of NPM1 in the spatial organization of nucleolus-associated heterochromatin.
Keywords: DNA Methyltransferase; Heterochromatin; Histone; Histone Chaperone; Histone Modification; Nucleolus; Ribosome Assembly; p53.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- McStay B., Grummt I. (2008) The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24, 131–157 - PubMed
-
- Hamperl S., Wittner M., Babl V., Perez-Fernandez J., Tschochner H., Griesenbeck J. (2013) Chromatin states at ribosomal DNA loci. Biochim. Biophys. Acta 1829, 405–417 - PubMed
-
- Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell. Biol. 8, 574–585 - PubMed
-
- Németh A., Längst G. (2011) Genome organization in and around the nucleolus. Trends Genet. 27, 149–156 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

