A circadian gene expression atlas in mammals: implications for biology and medicine
- PMID: 25349387
- PMCID: PMC4234565
- DOI: 10.1073/pnas.1408886111
A circadian gene expression atlas in mammals: implications for biology and medicine
Abstract
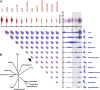
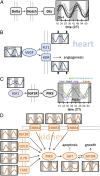
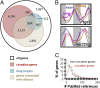
To characterize the role of the circadian clock in mouse physiology and behavior, we used RNA-seq and DNA arrays to quantify the transcriptomes of 12 mouse organs over time. We found 43% of all protein coding genes showed circadian rhythms in transcription somewhere in the body, largely in an organ-specific manner. In most organs, we noticed the expression of many oscillating genes peaked during transcriptional "rush hours" preceding dawn and dusk. Looking at the genomic landscape of rhythmic genes, we saw that they clustered together, were longer, and had more spliceforms than nonoscillating genes. Systems-level analysis revealed intricate rhythmic orchestration of gene pathways throughout the body. We also found oscillations in the expression of more than 1,000 known and novel noncoding RNAs (ncRNAs). Supporting their potential role in mediating clock function, ncRNAs conserved between mouse and human showed rhythmic expression in similar proportions as protein coding genes. Importantly, we also found that the majority of best-selling drugs and World Health Organization essential medicines directly target the products of rhythmic genes. Many of these drugs have short half-lives and may benefit from timed dosage. In sum, this study highlights critical, systemic, and surprising roles of the mammalian circadian clock and provides a blueprint for advancement in chronotherapy.
Keywords: chronotherapy; circadian; gene networks; genomics; noncoding RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Prevalence of cycling genes and drug targets calls for prospective chronotherapeutics.Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):15869-70. doi: 10.1073/pnas.1418570111. Epub 2014 Nov 3. Proc Natl Acad Sci U S A. 2014. PMID: 25368193 Free PMC article. No abstract available.
-
Circadian genetics: Timing is everything.Nat Rev Genet. 2014 Dec;15(12):780. doi: 10.1038/nrg3864. Epub 2014 Nov 11. Nat Rev Genet. 2014. PMID: 25385128 No abstract available.
-
Circadian Rhythms. Temporal targets of drug action.Science. 2014 Nov 21;346(6212):921-2. doi: 10.1126/science.aaa2285. Science. 2014. PMID: 25414292 No abstract available.
References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

