Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization
- PMID: 25349421
- PMCID: PMC4234544
- DOI: 10.1073/pnas.1419030111
Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization
Abstract
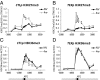
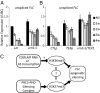
Long noncoding RNAs (lncRNAs) have been proposed to play important roles in gene regulation. However, their importance in epigenetic silencing and how specificity is determined remain controversial. We have investigated the cold-induced epigenetic switching mechanism involved in the silencing of Arabidopsis thaliana Flowering Locus C (FLC), which occurs during vernalization. Antisense transcripts, collectively named COOLAIR, are induced by prolonged cold before the major accumulation of histone 3 lysine 27 trimethylation (H3K27me3), characteristic of Polycomb silencing. We have found that COOLAIR is physically associated with the FLC locus and accelerates transcriptional shutdown of FLC during cold exposure. Removal of COOLAIR disrupted the synchronized replacement of H3K36 methylation with H3K27me3 at the intragenic FLC nucleation site during the cold. Consistently, genetic analysis showed COOLAIR and Polycomb complexes work independently in the cold-dependent silencing of FLC. Our data reveal a role for lncRNA in the coordinated switching of chromatin states that occurs during epigenetic regulation.
Keywords: Arabidopsis; Polycomb; flowering; histone modifications; long noncoding RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gendall AR, Levy YY, Wilson A, Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107(4):525–535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

