TALEN and CRISPR/Cas Genome Editing Systems: Tools of Discovery
- PMID: 25349712
- PMCID: PMC4207558
TALEN and CRISPR/Cas Genome Editing Systems: Tools of Discovery
Abstract
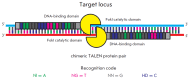
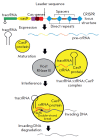
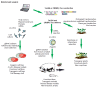
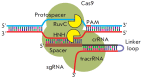
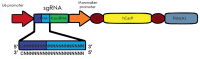
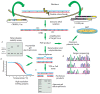
Precise studies of plant, animal and human genomes enable remarkable opportunities of obtained data application in biotechnology and medicine. However, knowing nucleotide sequences isn't enough for understanding of particular genomic elements functional relationship and their role in phenotype formation and disease pathogenesis. In post-genomic era methods allowing genomic DNA sequences manipulation, visualization and regulation of gene expression are rapidly evolving. Though, there are few methods, that meet high standards of efficiency, safety and accessibility for a wide range of researchers. In 2011 and 2013 novel methods of genome editing appeared - this are TALEN (Transcription Activator-Like Effector Nucleases) and CRISPR (Clustered Regulatory Interspaced Short Palindromic Repeats)/Cas9 systems. Although TALEN and CRISPR/Cas9 appeared recently, these systems have proved to be effective and reliable tools for genome engineering. Here we generally review application of these systems for genome editing in conventional model objects of current biology, functional genome screening, cell-based human hereditary disease modeling, epigenome studies and visualization of cellular processes. Additionally, we review general strategies for designing TALEN and CRISPR/Cas9 and analyzing their activity. We also discuss some obstacles researcher can face using these genome editing tools.
Keywords: CRISPR/Cas9; TALEN; genome editing.
Figures

Similar articles
-
Gene Editing With TALEN and CRISPR/Cas in Rice.Prog Mol Biol Transl Sci. 2017;149:81-98. doi: 10.1016/bs.pmbts.2017.04.006. Epub 2017 May 24. Prog Mol Biol Transl Sci. 2017. PMID: 28712502 Review.
-
Advances in Plant Epigenome Editing Research and Its Application in Plants.Int J Mol Sci. 2023 Feb 8;24(4):3442. doi: 10.3390/ijms24043442. Int J Mol Sci. 2023. PMID: 36834852 Free PMC article. Review.
-
CRISPR-Cas9 system: A new-fangled dawn in gene editing.Life Sci. 2019 Sep 1;232:116636. doi: 10.1016/j.lfs.2019.116636. Epub 2019 Jul 8. Life Sci. 2019. PMID: 31295471 Review.
-
Comparison of gene editing efficiencies of CRISPR/Cas9 and TALEN for generation of MSTN knock-out cashmere goats.Theriogenology. 2019 Jul 1;132:1-11. doi: 10.1016/j.theriogenology.2019.03.029. Epub 2019 Apr 1. Theriogenology. 2019. PMID: 30981084
-
CRISPR/Cas9: an advanced tool for editing plant genomes.Transgenic Res. 2016 Oct;25(5):561-73. doi: 10.1007/s11248-016-9953-5. Epub 2016 Mar 24. Transgenic Res. 2016. PMID: 27012546 Review.
Cited by
-
In Vivo Genome Editing as a Therapeutic Approach.Int J Mol Sci. 2018 Sep 12;19(9):2721. doi: 10.3390/ijms19092721. Int J Mol Sci. 2018. PMID: 30213032 Free PMC article. Review.
-
Can Designer Indels Be Tailored by Gene Editing?: Can Indels Be Customized?Bioessays. 2019 Dec;41(12):e1900126. doi: 10.1002/bies.201900126. Epub 2019 Nov 6. Bioessays. 2019. PMID: 31693213 Free PMC article.
-
Xenbase: a genomic, epigenomic and transcriptomic model organism database.Nucleic Acids Res. 2018 Jan 4;46(D1):D861-D868. doi: 10.1093/nar/gkx936. Nucleic Acids Res. 2018. PMID: 29059324 Free PMC article.
-
Reverse Genetics and High Throughput Sequencing Methodologies for Plant Functional Genomics.Curr Genomics. 2016 Dec;17(6):460-475. doi: 10.2174/1389202917666160520102827. Curr Genomics. 2016. PMID: 28217003 Free PMC article.
-
Proteome alterations during clonal isolation of established human pancreatic cancer cell lines.Cell Mol Life Sci. 2022 Oct 22;79(11):561. doi: 10.1007/s00018-022-04584-9. Cell Mol Life Sci. 2022. PMID: 36271971 Free PMC article.
References
-
- Capecchi M.R.. Nat. Rev. Genet. 2005;6(6):507–512. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
