Cloning and functional characterization of a vacuolar Na+/H+ antiporter gene from mungbean (VrNHX1) and its ectopic expression enhanced salt tolerance in Arabidopsis thaliana
- PMID: 25350285
- PMCID: PMC4211658
- DOI: 10.1371/journal.pone.0106678
Cloning and functional characterization of a vacuolar Na+/H+ antiporter gene from mungbean (VrNHX1) and its ectopic expression enhanced salt tolerance in Arabidopsis thaliana
Abstract
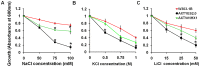
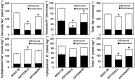
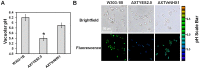
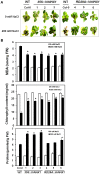
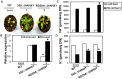
Plant vacuolar NHX exchangers play a significant role in adaption to salt stress by compartmentalizing excess cytosolic Na+ into vacuoles and maintaining cellular homeostasis and ionic equilibrium. We cloned an orthologue of the vacuolar Na+/H+ antiporter gene, VrNHX1 from mungbean (Vigna radiata), an important Asiatic grain legume. The VrNHX1 (Genbank Accession number JN656211.1) contains 2095 nucleotides with an open reading frame of 1629 nucleotides encoding a predicted protein of 542 amino acids with a deduced molecular mass of 59.6 kDa. The consensus amiloride binding motif (84LFFIYLLPPI93) was observed in the third putative transmembrane domain of VrNHX1. Bioinformatic and phylogenetic analysis clearly suggested that VrNHX1 had high similarity to those of orthologs belonging to Class-I clade of plant NHX exchangers in leguminous crops. VrNHX1 could be strongly induced by salt stress in mungbean as the expression in roots significantly increased in presence of 200 mM NaCl with concomitant accumulation of total [Na+]. Induction of VrNHX1 was also observed under cold and dehydration stress, indicating a possible cross talk between various abiotic stresses. Heterologous expression in salt sensitive yeast mutant AXT3 complemented for the loss of yeast vacuolar NHX1 under NaCl, KCl and LiCl stress indicating that VrNHX1 was the orthologue of ScNHX1. Further, AXT3 cells expressing VrNHX1 survived under low pH environment and displayed vacuolar alkalinization analyzed using pH sensitive fluorescent dye BCECF-AM. The constitutive and stress inducible expression of VrNHX1 resulted in enhanced salt tolerance in transgenic Arabidopsis thaliana lines. Our work suggested that VrNHX1 was a salt tolerance determinant in mungbean.
Conflict of interest statement
Figures




References
-
- Kronzucker HJ, Britto DT (2011) Sodium transport in plants: a critical review. New Phytol 189: 54–8. - PubMed
-
- Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444: 139–158. - PubMed
-
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant Cellular and Molecular Responses to High Salinity. Annu Rev Plant Physiol Plant Mol Bio 51: 463–99. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

