Development of an enhanced metaproteomic approach for deepening the microbiome characterization of the human infant gut
- PMID: 25350865
- PMCID: PMC4286196
- DOI: 10.1021/pr500936p
Development of an enhanced metaproteomic approach for deepening the microbiome characterization of the human infant gut
Abstract
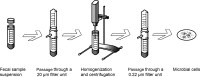
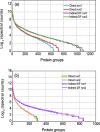
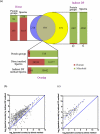
The establishment of early life microbiota in the human infant gut is highly variable and plays a crucial role in host nutrient availability/uptake and maturation of immunity. Although high-performance mass spectrometry (MS)-based metaproteomics is a powerful method for the functional characterization of complex microbial communities, the acquisition of comprehensive metaproteomic information in human fecal samples is inhibited by the presence of abundant human proteins. To alleviate this restriction, we have designed a novel metaproteomic strategy based on double filtering (DF) the raw samples, a method that fractionates microbial from human cells to enhance microbial protein identification and characterization in complex fecal samples from healthy premature infants. This method dramatically improved the overall depth of infant gut proteome measurement, with an increase in the number of identified low-abundance proteins and a greater than 2-fold improvement in microbial protein identification and quantification. This enhancement of proteome measurement depth enabled a more extensive microbiome comparison between infants by not only increasing the confidence of identified microbial functional categories but also revealing previously undetected categories.
Keywords: Metaproteome; double filtering; human infant gut; shotgun proteomics.
Figures


References
-
- Nell S.; Suerbaum S.; Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat. Rev. Microbiol. 2010, 8, 564–77. - PubMed
-
- Sanz Y.; Santacruz A.; Gauffin P. Gut microbiota in obesity and metabolic disorders. Proc. Nutr. Soc. 2010, 69, 434–41. - PubMed
-
- Berer K.; Mues M.; Koutrolos M.; Rasbi Z. A.; Boziki M.; Johner C.; Wekerle H.; Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011, 479, 538–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

