Leveraging the new with the old: providing a framework for the integration of historic microarray studies with next generation sequencing
- PMID: 25350881
- PMCID: PMC4251047
- DOI: 10.1186/1471-2105-15-S11-S3
Leveraging the new with the old: providing a framework for the integration of historic microarray studies with next generation sequencing
Abstract
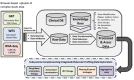
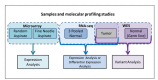
Next Generation Sequencing (NGS) methods are rapidly providing remarkable advances in our ability to study the molecular profiles of human cancers. However, the scientific discovery offered by NGS also includes challenges concerning the interpretation of large and non-trivial experimental results. This task is potentially further complicated when a multitude of molecular profiling modalities are available, with the goal of a more integrative and comprehensive analysis of the cancer biology.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

