UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter
- PMID: 25351492
- PMCID: PMC4247584
- DOI: 10.1105/tpc.114.130716
UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter
Abstract
In plants subjected to UV-B radiation, responses are activated that minimize damage caused by UV-B. The bZIP transcription factor ELONGATED HYPOCOTYL5 (HY5) acts downstream of the UV-B photoreceptor UV RESISTANCE LOCUS8 (UVR8) and promotes UV-B-induced photomorphogenesis and acclimation. Expression of HY5 is induced by UV-B; however, the transcription factor(s) that regulate HY5 transcription in response to UV-B and the impact of UV-B on the association of HY5 with its target promoters are currently unclear. Here, we show that HY5 binding to the promoters of UV-B-responsive genes is enhanced by UV-B in a UVR8-dependent manner in Arabidopsis thaliana. In agreement, overexpression of REPRESSOR OF UV-B PHOTOMORPHOGENESIS2, a negative regulator of UVR8 function, blocks UV-B-responsive HY5 enrichment at target promoters. Moreover, we have identified a T/G-box in the HY5 promoter that is required for its UV-B responsiveness. We show that HY5 and its homolog HYH bind to the T/G(HY5)-box cis-acting element and that they act redundantly in the induction of HY5 expression upon UV-B exposure. Therefore, HY5 is enriched at target promoters in response to UV-B in a UVR8 photoreceptor-dependent manner, and HY5 and HYH interact directly with a T/G-box cis-acting element of the HY5 promoter, mediating the transcriptional activation of HY5 in response to UV-B.
© 2014 American Society of Plant Biologists. All rights reserved.
Figures
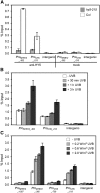

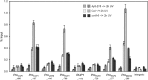
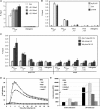
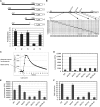
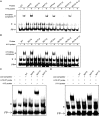
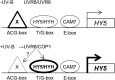
Comment in
-
How ELONGATED HYPOCOTYL5 helps protect plants from UV-B rays.Plant Cell. 2014 Oct;26(10):3826. doi: 10.1105/tpc.114.133363. Epub 2014 Oct 28. Plant Cell. 2014. PMID: 25351490 Free PMC article. No abstract available.
References
-
- Andronis C., Barak S., Knowles S.M., Sugano S., Tobin E.M. (2008). The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant 1: 58–67. - PubMed
-
- Ballare C.L., Barnes P.W., Flint S.D. (1995). Inhibition of hypocotyl elongation by ultraviolet-B radiation in de-etiolating tomato seedlings. I. The photoreceptor. Physiol. Plant. 93: 584–592.
-
- Beggs C.J., Wellmann E. (1994). Photocontrol of flavonoid biosynthesis. In Photomorphogenesis in Plants, 2nd ed, Kendrick R.E., Kronenberg G.H.M., eds (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 733–751.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

