Promoter-based integration in plant defense regulation
- PMID: 25352272
- PMCID: PMC4256871
- DOI: 10.1104/pp.114.248716
Promoter-based integration in plant defense regulation
Abstract
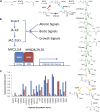
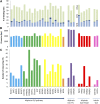
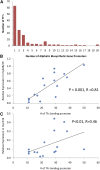
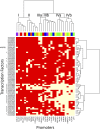
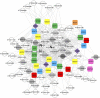
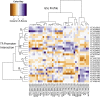
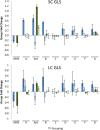
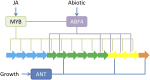
A key unanswered question in plant biology is how a plant regulates metabolism to maximize performance across an array of biotic and abiotic environmental stresses. In this study, we addressed the potential breadth of transcriptional regulation that can alter accumulation of the defensive glucosinolate metabolites in Arabidopsis (Arabidopsis thaliana). A systematic yeast one-hybrid study was used to identify hundreds of unique potential regulatory interactions with a nearly complete complement of 21 promoters for the aliphatic glucosinolate pathway. Conducting high-throughput phenotypic validation, we showed that >75% of tested transcription factor (TF) mutants significantly altered the accumulation of the defensive glucosinolates. These glucosinolate phenotypes were conditional upon the environment and tissue type, suggesting that these TFs may allow the plant to tune its defenses to the local environment. Furthermore, the pattern of TF/promoter interactions could partially explain mutant phenotypes. This work shows that defense chemistry within Arabidopsis has a highly intricate transcriptional regulatory system that may allow for the optimization of defense metabolite accumulation across a broad array of environments.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Alon U. (2006) An Introduction to Systems Biology: Design Principles of Biological Circuits, CRC Press, Taylor & Francis Group, Boca Raton, FL
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide Insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Asahina M, Azuma K, Pitaksaringkarn W, Yamazaki T, Mitsuda N, Ohme-Takagi M, Yamaguchi S, Kamiya Y, Okada K, Nishimura T, et al. (2011) Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc Natl Acad Sci USA 108: 16128–16132 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

