A cytoplasmic RNA virus generates functional viral small RNAs and regulates viral IRES activity in mammalian cells
- PMID: 25352551
- PMCID: PMC4227785
- DOI: 10.1093/nar/gku952
A cytoplasmic RNA virus generates functional viral small RNAs and regulates viral IRES activity in mammalian cells
Abstract
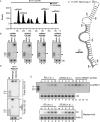
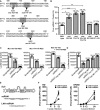
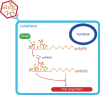
The roles of virus-derived small RNAs (vsRNAs) have been studied in plants and insects. However, the generation and function of small RNAs from cytoplasmic RNA viruses in mammalian cells remain unexplored. This study describes four vsRNAs that were detected in enterovirus 71-infected cells using next-generation sequencing and northern blots. Viral infection produced substantial levels (>10(5) copy numbers per cell) of vsRNA1, one of the four vsRNAs. We also demonstrated that Dicer is involved in vsRNA1 generation in infected cells. vsRNA1 overexpression inhibited viral translation and internal ribosomal entry site (IRES) activity in infected cells. Conversely, blocking vsRNA1 enhanced viral yield and viral protein synthesis. We also present evidence that vsRNA1 targets stem-loop II of the viral 5' untranslated region and inhibits the activity of the IRES through this sequence-specific targeting. Our study demonstrates the ability of a cytoplasmic RNA virus to generate functional vsRNA in mammalian cells. In addition, we also demonstrate a potential novel mechanism for a positive-stranded RNA virus to regulate viral translation: generating a vsRNA that targets the IRES.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. - PubMed
-
- Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. - PubMed
-
- Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. - PubMed
-
- Cullen B.R. Viruses and microRNAs. Nat. Genet. 2006;38(Suppl):S25–S30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

