Characterization of the rainbow trout spleen transcriptome and identification of immune-related genes
- PMID: 25352861
- PMCID: PMC4196580
- DOI: 10.3389/fgene.2014.00348
Characterization of the rainbow trout spleen transcriptome and identification of immune-related genes
Abstract
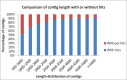
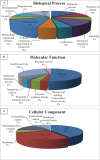
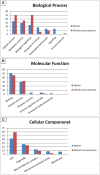
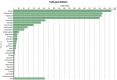
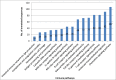
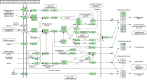
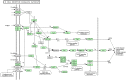
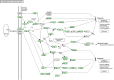
Resistance against diseases affects profitability of rainbow trout. Limited information is available about functions and mechanisms of teleost immune pathways. Immunogenomics provides powerful tools to determine disease resistance genes/gene pathways and develop genetic markers for genomic selection. RNA-Seq sequencing of the rainbow trout spleen yielded 93,532,200 reads (100 bp). High quality reads were assembled into 43,047 contigs. 26,333 (61.17%) of the contigs had hits to the NR protein database and 7024 (16.32%) had hits to the KEGG database. Gene ontology showed significant percentages of transcripts assigned to binding (51%), signaling (7%), response to stimuli (9%) and receptor activity (4%) suggesting existence of many immune-related genes. KEGG annotation revealed 2825 sequences belonging to "organismal systems" with the highest number of sequences, 842 (29.81%), assigned to immune system. A number of sequences were identified for the first time in rainbow trout belonging to Toll-like receptor signaling (35), B cell receptor signaling pathway (44), T cell receptor signaling pathway (56), chemokine signaling pathway (73), Fc gamma R-mediated phagocytosis (52), leukocyte transendothelial migration (60) and NK cell mediated cytotoxicity (42). In addition, 51 transcripts were identified as spleen-specific genes. The list includes 277 full-length cDNAs. The presence of a large number of immune-related genes and pathways similar to other vertebrates suggests that innate and adaptive immunity in fish are conserved. This study provides deep-sequence data of rainbow trout spleen transcriptome and identifies many new immune-related genes and full-length cDNAs. This data will help identify allelic variations suitable for genomic selection and genetic manipulation in aquaculture.
Keywords: KEGG; annotation; full-length cDNA; immune-related genes; spleen transcriptome; spleen-specific genes.
Figures


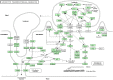

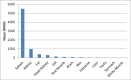
References
-
- Allendorf F. W., Thorgaard G. H. (1984). Tetraploidy and the evolution of salmonid fishes, in Evolutionary Genetics of Fishes, ed Turner B. J. (New York, NY: Plenum Press; ), 1–53
LinkOut - more resources
Full Text Sources
Other Literature Sources

