Genome-scale quantitative characterization of bacterial protein localization dynamics throughout the cell cycle
- PMID: 25353361
- PMCID: PMC4309519
- DOI: 10.1111/mmi.12841
Genome-scale quantitative characterization of bacterial protein localization dynamics throughout the cell cycle
Abstract
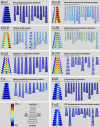
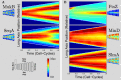
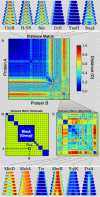
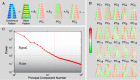
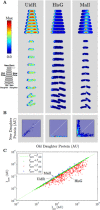
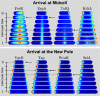
Bacterial cells display both spatial and temporal organization, and this complex structure is known to play a central role in cellular function. Although nearly one-fifth of all proteins in Escherichia coli localize to specific subcellular locations, fundamental questions remain about how cellular-scale structure is encoded at the level of molecular-scale interactions. One significant limitation to our understanding is that the localization behavior of only a small subset of proteins has been characterized in detail. As an essential step toward a global model of protein localization in bacteria, we capture and quantitatively analyze spatial and temporal protein localization patterns throughout the cell cycle for nearly every protein in E. coli that exhibits nondiffuse localization. This genome-scale analysis reveals significant complexity in patterning, notably in the behavior of DNA-binding proteins. Complete cell-cycle imaging also facilitates analysis of protein partitioning to daughter cells at division, revealing a broad and robust assortment of asymmetric partitioning behaviors.
© 2014 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures

References
-
- Alley MR, Maddock JR. Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. - PubMed
-
- Baneyx F. Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. - PubMed
-
- Bobola N, Jansen RP, Shin TH. Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

